The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
This paper argues that the issue with the origin of life and the origin of biodiversity is not an issue of time, though estimations of deep time are problematical. Instead the issue is the unbridgeable discontinuities among different life forms, discontinuities between biological taxa that we believe cannot be crossed by the mechanisms of random variations and natural selection. These discontinuities present themselves, time and again, as unmovable walls during molecular cloning. In an attempt to integrate the observations of fossils, studies of functions of genes and gene contents of various genomes, and lessons learned from molecular cloning, a new model designed to more accurately reflect the relationships between living things on earth is provided. Our hope is that this model will facilitate the functional annotation of genomes and the classification of organisms.
Keywords: convergent evolution, horizontal or lateral gene transfer, taxonomically restricted genes, tree of life, origin of life, origin of biodiversity, age of the universe, age of the earth
Disclaimer: The opinions expressed in this article are the author’s own and not necessarily those of the University of Missouri.
Introduction
The most fundamental questions in life are: What is life? Where did life come from? How are different life forms related? Eldredge and Gould (1972) spent about one third of their paper entitled “Punctuated equilibria: an alternative to phyletic gradualism” talking about how an individual’s presuppositions determine his or her interpretation of data or observations. They claimed, quite correctly, that “The expectations of theory color perception to such a degree that new notions seldom arise from facts collected under the influence of old pictures of the world” (Eldredge and Gould 1972, 83). In the field of biological studies, there is an urgent need to escape from the suffocating Darwinian tree of life model. In this paper I will first provide evidences of discontinuities among categories of organisms. Then I will describe a new model of how different life forms are related. A brief discussion of problems associated with the most popular estimates of the age of the universe and the age of the earth can be found in the Appendix.
Discontinuities at the Organismal Level
This includes discontinuity in morphology, in reproduction, and in the fossil record. All have been noticed and analyzed by many scientists. For morphological comparisons and reproduction isolation (hybridization) experiments please refer to (Frair 2000; Lightner 2006, 2007, 2010, 2011, 2012; ReMine 1990; Robinson and Cavanaugh 1998; Sanders and Wise 2003; Tomkins 2013; Wise 1990; Wood 2006, 2013; Wood et al. 2003). In fact, it is the morphological discontinuities between organisms that allowed Carl Linnaeus to classify organisms into different taxa (Linnaeus 1758).
Fossil discontinuity, or the lack of transitional fossils—“the intermediate links,” was regarded by Darwin as “the most obvious and serious objection which can be urged against” his theory On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life (Darwin 1859). Darwin thought that this resulted from the incompleteness of fossil collection. To fill this supposed deficiency in fossil collections, scientists have been arduously searching and digging for the last 150 years. Reflecting on the fossil collections of others and their own, Eldredge and Gould (1972) concluded that the fossil collections were essentially complete and yet lacked the transitional fossils, that the norm for a species was stability, and that speciation was a rare and difficult event that punctuates a system in homeostatic equilibrium. They believed that the transitional forms were not found because of allopatric speciation where new species can arise only when a small local population becomes isolated at the margin of the geographic range of its parent species and that the speciation occurs rapidly. The new species, which has become unable to mate and reproduce with its parental species, later migrated back to where its ancestors once lived.
An alternative explanation, which is a logical conclusion of the new model of life to be explained below, is that the fossil discontinuity is a reflection of the discontinuity between different life forms and the transitional forms have not been found because they never actually existed. The truth is that what we can be sure of about a fossil is that the fossilized organism once lived and then died. If one insists on an age association of the fossils, he/she will be greeted by a history of morphological disparity preceding diversity and persistent morphological discontinuity, opposite to the prediction of Darwinian evolution (Eldredge and Gould 1972; Meyer, Ross, Nelson, and Chien 2003; Meyer 2013).
Discontinuities at the Molecular Level
Discontinuities revealed by distinctions of vital molecular machineries
This is demonstrated in the basic molecular machineries that are necessary for the survival and propagation of all organisms, specifically those for DNA replication, transcription, and translation. As detailed recently, the molecular machineries for DNA replication, transcription, and translation cannot be interchanged between prokaryotes and eukaryotes, creating multiple unbridgeable discontinuities between them (Tan and Tomkins 2015a,b).
Discontinuities revealed by taxonomically restricted distribution of essential genes
An essential gene is a gene in an organism that is necessary for the viability of the organism. An organism dies when any one of its essential genes or essential non-coding DNA elements does not function properly. Interestingly, the vast majority (78.6%) of bacterial essential genes are bacterial domain specific, and even a higher percentage (95.5%) of eukaryotic essential genes are eukaryotic domain specific (Tan 2015). The domain specific distribution of bacterial and eukaryotic essential genes makes it unlikely for prokaryotes and eukaryotes to have been derived from a common ancestor or to have shared a common phylogenetic tree. This is because of the very nature of the essential genes and of the essential non-coding DNA elements, namely the essential non-genes, of an organism, that the organism cannot live without all of its essential genes and essential non-genes, and because of the inability of mutation and natural selection to create novel genes. Furthermore, mutagenesis studies have shown that, of all the possible arrangements of amino acids, only an extremely tiny fraction can function as a natural protein, that no novel genes have been observed to result from spontaneous mutation and natural selection, that two beneficial mutations often cancel each other’s functions, and that for each constructive mutation there are many sidetracks that prevent an organism from taking the constructive path (Axe 2004; Gauger et al. 2010; Tan 2015; Taylor et al. 2001).
Not only are there unbridgeable discontinuities in between domains of life, but also within each domain of life, as shown by the taxonomically restricted distribution of the essential genes of diverse organisms, including seven bacteria (Mycoplasma genitalium, Bacillus subtilis, Helicobacter pylori, Haemophilus influenza, Acinetobacter, Escherichia coli, and Caulobacter crescentus) and five eukaryotes (Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Homo sapiens) (Tan 2015). Therefore, life forms on earth are best represented as a forest of family trees.
Discontinuities revealed by taxonomically restricted distribution of essential non-genes
These include differences of origins of DNA replication, special regions of genomes where DNA replication initiates, and differences of enhancers and promoters of genes, regions of DNA that determine whether, when, and how much a gene will be expressed (Tan 2015).
Discontinuities revealed by molecular cloning
The discontinuities revealed by comparative studies of DNA replication, transcription, and translation, as well as information with respect to essential genes and essential non-genes between different organisms, have also been shown by molecular cloning. In addition, molecular cloning has revealed multiple other discontinuities as shown below.
1. Necessity of Essential Genes
The first potential discontinuity revealed by molecular cloning is the availability of essential genes. The pinnacle of molecular cloning up until the present has been the generation of Mycoplasma mycoides JCVI-syn1.0 cells using Mycoplasma mycoides subspecies capri as the genome donor, and its close relative Mycoplasma capricolum subspecies capricolum as the recipient (Gibson et al. 2010). Strikingly, one single base pair deletion in dnaA, one of the essential genes of M. mycoides, is enough to reduce all cloning efforts to nothing; not a single colony made of the synthesized genome could be generated until the missed base was added back manually. This confirms the notion that indeed every essential gene in an organism is necessary for the viability of that organism.
A reasonable inference from the nature of essential genes is that an organism A cannot evolve into another organism B that contains organism B-specific essential genes, that is, genes that are necessary for the survival of organism B but do not have homologs in organism A. In other words, all essential genes of an organism must have homologs in its ancestor, though these homologs may not be necessary for the survival of the ancestor, because each B-specific essential gene will create an unbridgeable discontinuity between A and B.
2. Presence of Potential Toxic Gene Products
The second potential discontinuity revealed by molecular cloning, which is on the opposite end of the spectrum from essential genes for an organism, is the presence of potential toxic genes, those that bring death to its potential host organisms into which they might be cloned. When Itaya and colleagues tried to clone the photosynthetic bacterium Synechocystis PCC6803 genome into the mesophilic bacterium B. subtilis 168, two Synechocystis RNA operons, rrnA and rrnB, blocked them from achieving their goal (Itaya et al. 2005). Individually, neither rrnA nor rrnB, can kill, but together they are fatal, suggesting that the lethality may be caused by their translation product(s). In support of this hypothesis, a number of Synechocystis genes were transcribed but very few Synechocystis proteins were made in B. subtilis. In addition, the cells with the hybrid genome, which is made of the 3.5-megabase (Mb) genome of Synechocystis and the 4.2-Mb genome of the B. subtilis, still behave like a B. subtilis cell, growing only in the medium for culturing B. subtilis but not in the medium for culturing Synechocystis.
Later, when Karas and colleagues tried to clone the Acholeplasma laidlawii PG-8A genome in S. cerevisiae as a yeast centromeric plasmid, an A. laidlawii extracellular endonuclease acted as the lethal factor (Karas et al. 2012). Whenever this endonuclease is expressed in yeast, it kills the yeast host cells.
An elegant, systematic, and large-scale study of toxic genes has been performed by Sorek and colleagues (Sorek et al. 2007). They found that the main reason that some of the genomic regions of many organisms cannot be cloned in E. coli is that those regions produce toxic proteins in E. coli. They cloned 39 such genes into an E. coli inducible expression vector. In the absence of the expression inducer, the bacteria grew. In the presence of the inducer, 32 of the 39 genes (82%) inhibited E. coli growth. Furthermore, they found that similar genes from different organisms sometimes behaved differently. For example, ribosomal protein coding genes L4/L1e and S12 from some organisms cannot be cloned in E. coli, while L4/L1e and S12 genes from other organisms can be readily cloned in E. coli. They found this to be due to differences in the promoters of these genes. The promoters of those clonable L4/L1e and S12 genes are not recognized by the E. coli host. Consequently, they are not transcribed, or expressed, in E. coli and, thus, their coded proteins are not produced. When the clonable L4/L1e and S12 genes are cloned under the control of an E. coli inducible promoter they are able to kill the host cells when their expression is induced. Indeed, it is a general practice in cloning for expression to clone a potentially toxic gene under the control of an inducible promoter, which ensures that no harmful products will be made in the absence of the inducer so that the cloned DNA can be maintained and amplified in its host cells.
A surprising finding of Sorek and colleagues is the importance of gene dosage (Sorek et al. 2007). They found that some of the genes from E. coli HS resist being transferred into E. coli K12 cells, even though the E. coli HS genes have >98% identity to the E. coli K12 genes. This suggests that duplication of certain genes may be lethal to an organism, thus limiting the scope of gene duplication.
Several lessons can be learned here. First, the necessity of one organism may be the annihilator of another. The potential toxic effect of the gene products of one organism on another will create a boundary preventing intermingling of the two organisms. Second, a gene that cannot be expressed cannot do anything, bad or good; thus, this gene will be invisible to any natural selection. In fact, this gene will only be a metabolic burden to the host organism. Such unused genes tend to become degenerated or totally deleted, which is why cloned genomes will be lost without continued selection (Karas et al. 2012). Third, genes, including DNA, RNA, and proteins, are physical entities. They follow natural laws. As long as a DNA binding protein sees its binding target, it will bind—it will not think and it cannot think. When the E. coli transcription machinery sees a promoter that looks like an E. coli gene promoter, the transcription machinery will bind the promoter, transcribe the gene, and the E. coli translation machinery will translate the resultant transcript. If the transcript contains the necessary sequence to make a protein, it will make it, even if it kills the host E. coli cell. Both the transcription and the translation machineries will do that, predictably, repeatedly. They will behave as determined by their chemical and physical properties. They will not think because they cannot think. They will not choose because they are unable to tell whether a gene comes from the E. coli genome or from a foreign genome. A DNA segment with the sequence ATGC will be the same ATGC sequence whether it be from E. coli or from a different organism. In other words, a cell uses what is available and lives in the present. It does not care about the future. This limits what a cell can do. This should also limit one’s tendency to claim that “the bacteria or evolution somehow figured out a way to . . .”.
3. Distinction of Essential Non-Genes
The third potential discontinuity revealed by molecular cloning is the incompatibility of the essential non-genes, of which the best known is the origin of DNA replication. Even though not every person doing molecular cloning thinks about the origin of DNA replication—most people only care about the multiple cloning sites—the origin of replication is one of the most important features of a cloning vector. To clone a gene in E. coli, one has to use a vector that contains an origin of replication that functions as an origin of replication in E. coli. To clone a gene in S. cerevisiae, one has to use a vector that contains an origin of replication that functions as an origin of replication in S. cerevisiae. To clone a gene in a vector that can grow in both E. coli and S. cerevisiae one has to use a vector, known as a shuttle vector (Fig. 1), that contains an origin of replication that functions as an origin of replication in E. coli and another origin of replication that functions as an origin of replication in S. cerevisiae, because the origin of replication of E. coli and that of S. cerevisiae differ in their structures and differ in their ways being recognized and activated (Tan and Tomkins 2015b). This is part of the reason why DNA replication machineries are non-exchangeable between prokaryotes and eukaryotes, generating an unbridgeable discontinuity between them.
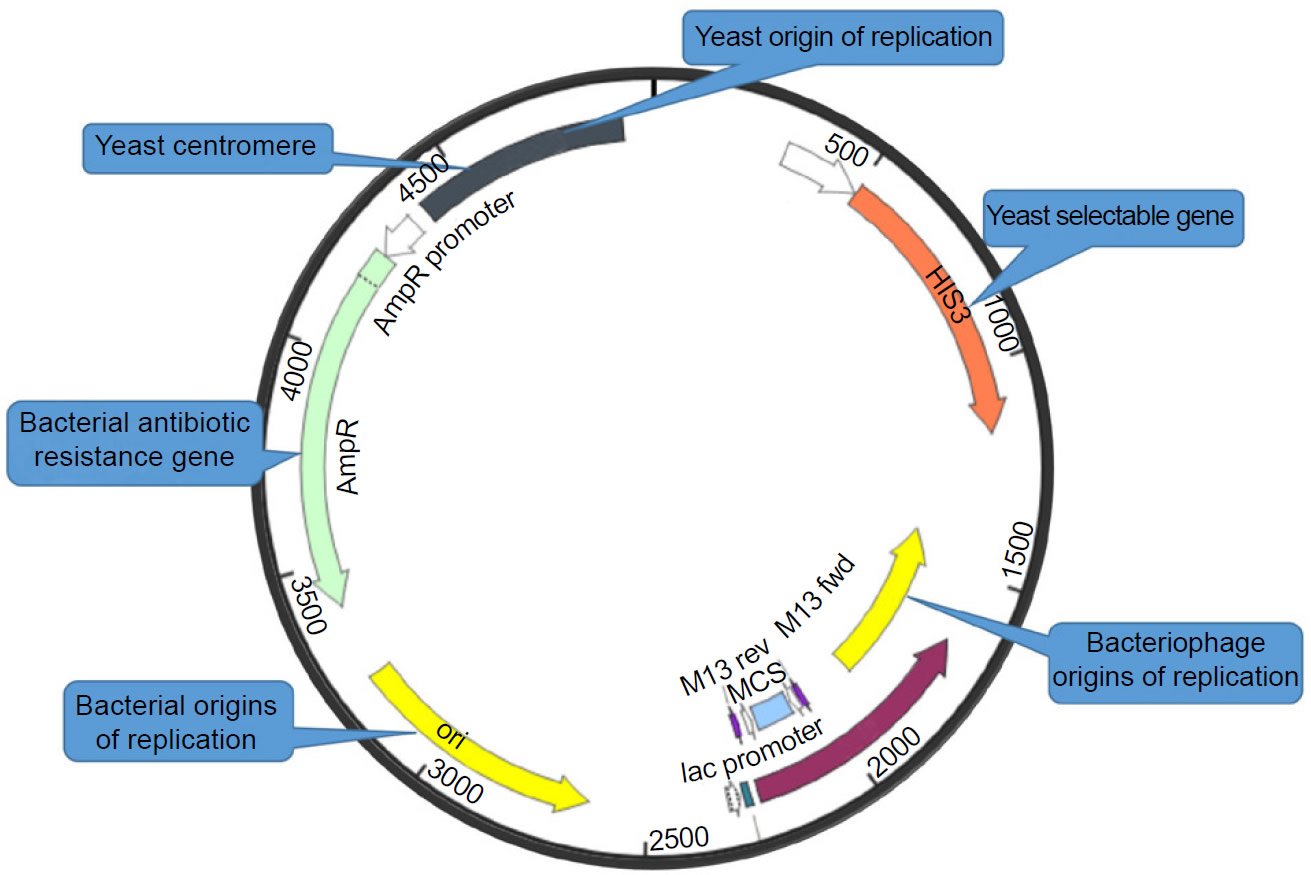
Fig. 1. An E.coli-yeast shuttle vector. Plasmid map is adapted from SnapGene (http://www.snapgene.com/resources/plasmid_files/yeast_plasmids/pRS313/). Used with permission.
Not only are the origins of replication from organisms as far from one another as E. coli, a prokaryote, and S. cerevisiae, a eukaryote, not interchangeable, the origins of replication from closely related organisms may not be exchangeable. For example, plasmids constructed for different species belonging to the Mycoplasma genus, including the genome donor M. mycoides subsp. mycoides large colony (MmmLC) and the genome receiver M. capricolum subsp. capricolum of M. mycoides JCVI-syn1.0, and three others: Spiroplasma citri, Mycoplasma pulmonis, M. mycoides subsp. mycoides small colony (MmmSC), can grow in their homologous hosts but may not grow in heterologous hosts (Lartigue et al. 2003). Of these five Mycoplasma species, M. capricolum is the broadest host; plasmids with the origins of replication (oriC) from all but M. pulmonis can be replicated in M. capricolum. In contrast, the plasmid with M. capricolum oriC cannot be replicated even in its closely related species MmmSC and MmmLC. This was the reason why Gibson and colleagues (2010) chose MmmLC as the genome donor and the M. capricolum as the receiver, not the reverse, to create their first artificial bacterium. Furthermore, plasmids with M. pulmonis oriC can only grow in M. pulmonis.
Interestingly, not only the identity but also the number of the origins of replication is important when cloning long pieces of DNA. For instance, when cloning the genomic DNA of Synechococcus elongatus PCC 79422767, a high G+C bacterium (G/C content 55%), in S. cerevisiae, insertion of yeast origins of replication throughout the S. elongatus DNA is necessary to clone and maintain S. elongatus DNA fragments over 200 kb (Noskov et al. 2012).
Therefore, the differences in the origins of DNA replication between different organisms, sometimes even between very closely related ones, can be a potent generator of discontinuities between different life forms, even their sharing of plasmids.
4. Differences of Promoters
The fourth potential generator of discontinuities revealed by molecular cloning is the differences among promoters. To express a gene in vitro or in a living host organism, one has to clone the gene into a vector under the control of a promoter recognizable by the organism or the expression system. To express a gene in E. coli, one has to clone the gene in an E. coli vector and under the control of a promoter that can be recognized by the E. coli transcription machinery. To express a gene in S. cerevisiae, one has to clone the gene in a S. cerevisiae vector and under the control of a promoter that can be recognized by the S. cerevisiae transcription machinery. Not surprisingly, in the E. coli-yeast shuttle vector shown in Fig. 1, the yeast selectable marker gene HIS is under the control of a yeast promoter, while the bacterial selectable marker genes, the antibiotic resistance gene and the lacZ gene, are under the control of bacterial promoters (Fig. 1). This kind of arrangement is necessary because the promoters of E. coli and those of S. cerevisiae differ in their structures and are recognized by different proteins; they are not interchangeable (Tan and Tomkins 2015b). This is another evidence of the fundamental discontinuities between life forms.
5. Presence of Restriction Systems
The fifth potential generator of discontinuities revealed by molecular cloning is the presence of restriction systems, in that an organism may contain restriction enzymes that will cut foreign DNA introduced into its cell. This is another hurdle that the Venter group had to overcome when they made their M. mycoides JCVI-syn1.0 cells (Gibson et al. 2010; Lartigue et al. 2009). It turned out that the M. capricolum is able to take and maintain the M. mycoides genomic DNA made in M. mycoides cells, but it will digest the M. mycoides genomic DNA made in yeast because the endogenous M. mycoides genomic DNA is methylated, while yeast lacks the enzymes to methylate the M. mycoides genomic DNA properly. The required methyltransferases that are encoded by the M. mycoides JCVI-syn1.0 genomic DNA cannot be made by yeast because their promoters cannot be recognized by the yeast transcription machinery and because the codon incompatibility between yeast and M. mycoides that will be discussed below. Therefore, the Venter team had to either methylate the synthetic DNA in vitro with extracts from M. capricolum or from M. mycoides or with purified M. mycoides methyltransferases, or, alternatively, to inactivate the M. capricolum restriction enzyme. Oddly, since M. capricolum and M. mycoides share the same restriction system, the yeast-grown M. mycoides genomic DNA will not survive its own restriction system, and not be welcomed back to its biological parent’s home, not until it is wrapped with the armor of proper methylation.
6. Incompatibility of DNA Composition
The sixth potential generator of discontinuities revealed by molecular cloning is the DNA composition, that is, the G/C contents of genomes, especially when the G/C content of the genome to be cloned differ too much from that of the host genome. For example, cloning the abovementioned S. elongatus PCC 7942 genome, which has a G/C content of 55%, in yeast, which has a G/C content of 38%, was difficult; S. elongatus DNA fragments over ~200 kb were not stably maintained when only one yeast origin of replication was used (Noskov et al. 2012). Only after addition of yeast origins of replication throughout, at intervals of <150 kb, S. elongatus DNA fragments of 454 kb could be cloned in yeast. On the other hand, genomes with G/C content (32% to 38%) similar to yeast, as big as 1.6 Mb, have been cloned in yeast with one yeast autonomously replicating sequences (ARS) from the yeast cloning vector (Gibson et al. 2008, 2010; Tagwerker et al. 2012).
7. Incompatibility of Genetic Codons
The seventh potential generator of discontinuities revealed by molecular cloning is codon incompatibility, including differences in the meanings of codons and differences in the frequency of codon usage between two organisms. For example, UGA codes a tryptophan in M. mycoides, while in yeast and most other organisms it does not code for any amino acid but serves as a translation stop signal. This is like a double-edged sword. It is advantageous when one wishes to clone the M. mycoides genomic DNA in yeast because the M. mycoides proteins may be truncated, even if its genes are driven by cryptic promoters active in yeast, so that no toxic proteins will be generated (Lartigue et al. 2009). However, to express an M. mycoides protein that contains tryptophan codons in yeast will not be possible unless the M. mycoides tryptophan codons are changed to the yeast tryptophan codons.
Codon usage frequency, or codon biases, can become the bottleneck in molecular cloning, especially when one’s purpose is to produce high levels of proteins. This is clearly demonstrated with the expression of the fragment C domain of the tetanus toxin (TetC) from Clostridium tetani in various host cells. In E. coli, removing the E. coli rare codons allowed the expression level to increase from 2~3% to ~14% of cell proteins (Makoff et al. 1989). In S. cerevisiae, no TetC can be generated with the original TetC gene, while the codon-modified TetC gene, with a high-GC content, was expressed to 2~3% of soluble cell proteins (Romanos et al. 1991). In Pichia pastoris, a different yeast strain, the high-GC TetC can be expressed up to 27% of total cell proteins (Clare, Sreekrishna, and Romanos 1998). In tobacco chloroplasts, both the original low-GC and the synthetic high-GC TetC genes are expressed at high levels (Tregoning et al. 2003). Of course, in each of the expression systems, the TetC gene has to be cloned into a vector that contains an origin of replication and under the control of a promoter designed for the specific expression host organism.
Therefore, molecular cloning has revealed the existence of various potential discontinuities between organisms. These discontinuities highly constrain the number of changes that will result in a viable life form, thus generating gaps that are unbridgeable via natural means, although these barriers can be overcome by human intelligence and engineering.
The Two Sides of the Same Coin
In addition to discontinuity, another characteristic shared by various life forms is similarity. These two—similarity and discontinuity—constitute the two sides of the same coin of every organism when it is compared with other life forms.
As is the case with discontinuities, similarities can also be at the molecular level or the organismal level. On the molecular level, we have the basic building blocks of DNA, RNA, and proteins; the genetic code; the necessity and the chemistry of DNA replication, transcription, and translation; similarity of sequences and/or structures of molecules performing similar functions in different organisms; etc. On the organismal level, the numbers and the organization of bones in the limbs of vertebrates, as well as the texture and function of skin, heart, liver, or kidney tissues of various animals, etc.
Similarity is normally referred to as homology and is generally considered to be the result of descent from a common ancestor. However, it is equally consistent with creation by a common designer. Admittedly, homology-from-common-ancestor is simpler than homology-by-common-designer if different life forms could have evolved from a common ancestor naturally because it does not require an external agent—God in the natural world, man in the cloning experiments. However, the unbridgeable discontinuities between different organisms render their generation by natural means, for example, by mutations and natural selection, impossible.
A special showcase of the organic combination of similarity and discontinuity among living beings is mosaics, the combination of structures or genes of organisms showing discontinuity between closely related species, yet similarity between unrelated lineages. For example, both the little brown bat and an unrelated non-bat species, the bottlenose dolphin, use echolocation, while two other bat species, the straw-colored fruit bat and the large flying fox, do not (Parker et al. 2013).
In fact, all the genomes of bacteria, archaea, and eukaryotes are mosaics. Bacteria and archaea are similar in that they have neither nuclei, nor spliceosomal introns, nor spliceosomal intron splicing machinery, and in that transcription and translation occur in the same compartment, even using a transcript for translation while it is being synthesized. However, the membranes of bacteria are more similar to that of eukaryotes, but very distinct from those of archaea. On the other hand, the molecular machines involved in DNA replication, transcription, or translation, of archaea are very different from that of bacteria, but are similar to that of eukaryotes (Tan and Tomkins 2015a, b). Interestingly, although the DNA replication and transcription machineries of archaea are more similar to that of eukaryotes than that of bacteria, they are different enough to prevent those used by archaea from being used by eukaryotes, making it impossible for archaea, bacteria, or eukaryotes to have shared a common ancestor. This is a clear example of discontinuities within similarity.
Mosaics are a nuisance for the evolution model and are normally claimed to result from convergent evolution or lateral gene transfer. Convergent evolution is the alleged independent evolution of similar features or genes in species of different lineages. Lateral gene transfer is a process in which a gene of an organism is transferred to an unrelated organism in a way other than the vertical transfer from a parent to its offspring. It is worth pointing out that both convergent evolution and lateral gene transfer have been evoked to explain the phenomena of mosaics, not based on observation. Lateral gene transfer is often claimed to be the vehicle when a computer-generated phylogenetic tree does not fit the predictions of the evolution model.
In contrast, mosaics are characteristics of objects that have been designed by intelligent humans. For example, there are real American flags, some big and some small. There are also American flag shirts, pants, and shoes, not due to evolution but due to design (Fig. 2). These American flag-containing items can be organized according to their function or morphology, similarly to the way in which Haeckel organized different life forms (Dayrat 2003). Or, they can be organized based on their material makeups, as people build molecular phylogenetic trees according to the sequences of DNA, RNA, or proteins (Puigbò, Wolf, and Koonin 2009; Woese 2000; Woese and Fox 1977).
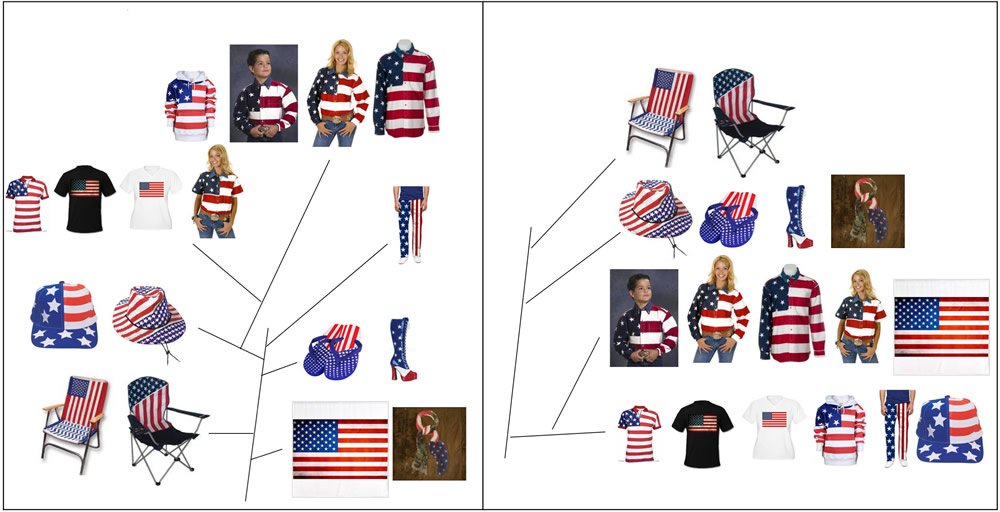
Fig. 2. Items with images of the American flags arranged by function (A) or by materials (B). All pictures are from Google search.
Similarity and discontinuity, as well as mosaics, are common phenomena in biology and need to be explained. Both the evolution model and the creation model can explain similarity. If an organism indeed can naturally evolve from another then evolution is a simpler choice, and thus, according to the principle of Ockham’s Razor, it would be the preferred mechanism. On the other hand, discontinuities and mosaics can be easily explained with the creation model, while evolution will work only if the claimed convergent evolution or lateral gene transfer indeed occurred. As discussed above, discontinuity is a death sentence for Darwinian evolution but a norm for intelligent designs.
Convergent evolution has been claimed to be responsible for the independent generation of similar traits (body structures, coloration, or organs)—traits that are lacking in their assumed ancestors—in unrelated or distantly related organisms in multiple lineages, including mammals, reptiles, fish, birds, arthropods, and mollusks. For example, koalas of Australasia have fingerprints indistinguishable from those of humans, and some whales use echolocation like some bats (https://en.wikipedia.org/wiki/List_of_examples_of_convergent_evolution, List of examples of convergent evolution).
Convergent evolution is unlikely to happen because it requires the simultaneous generation of multiple new genes—hundreds in the case of the echolocation in bats and whales (Parker et al. 2013), but gene generation via mutation and natural selection is improbable (Tan 2015). In addition, some of the genes functioning in the processes that are mosaic are taxonomically-restricted essential genes whose function is indispensable for the survival of its carrier organism (Tan 2015, and references therein).
Why do organisms on earth show not only similarity but also discontinuity, two characters seemingly contradictory to one another? Could it be that many biological processes or molecules are designed to be similar enough so that we can study them and can apply the knowledge gained from studying one organism to another, yet they are different enough to demonstrate that the mosaics are products of intelligent designer instead of products of evolution through accumulated mutations and natural selection?
A New Model of the Relationship of Different Types of Organisms
More and more cognitive observations of extant organisms, studies on hybridization, the fossil record, and characterization of genes and genomes are converging to support the conclusion that life forms on earth are best represented as a forest of family trees. This conclusion has been substantiated by many molecular cloning experiments. The next step is to define and characterize the forest, including its size and scope, what organisms belong to a specific family tree, how different trees are related, and how different branches of the same tree are related. It is conceivable that the process will not be easy, because as John Keynes said “The difficulty is not in generating new ideas but in escaping from the old ones” (quoted in Sverdlov 2006). To break loose from the stronghold of Darwinian evolution, escaping the paradigm of one family tree connecting all organisms, will not be an easy task because many people are absolutely convinced of the truth of the theory.
My working model is that all organisms on earth can be organized into a forest of family trees (Fig. 3). Branches on the same tree are all offspring either of one asexually reproducing parent, or of a population of ancestors that contain the same genes, or of a pair of sexually reproducing parents. Lines in between different family trees link characters (phenotypes, genes, or pathways) that are shared among organisms that do not belong to the same family trees. These characters include the many cases of alleged convergent evolution. I think such characters reflect the modular nature of life. In a nutshell, two key concepts, or hypotheses, or assumptions, in the model are that life is modular and that not all organisms belong to one family tree. Each organism is a combination of multiple modules. A module can be a signal transduction pathway, a biological process, or a special structure. For example, each of the common signal transduction pathways, including the Wnt pathway, the hedgehog pathway, the TGFß pathway, the Notch pathway, can be a module. Organisms on different family trees can share the same or similar modules. Each module may be tailored to individual trees. In fact, comparison of organisms on different family trees will be an efficient way to identify and characterize various modules, the more diverse the better. A definitive distinction concerning whether two organisms can belong to the same family tree is whether they share the same taxonomically restricted essential genes and essential non-genes, while a practical approximation is whether they share the same taxonomically restricted genes and non-genes (Tan 2015).
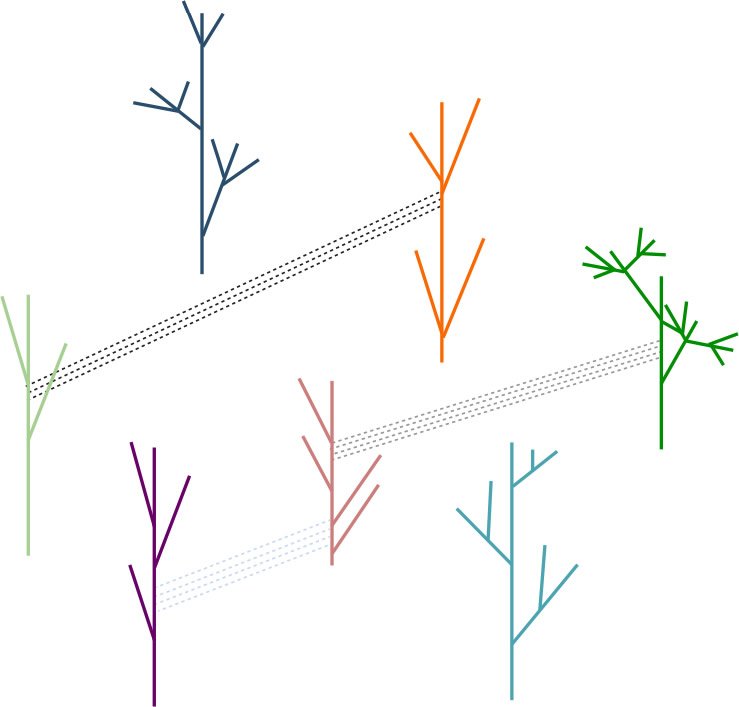
Fig. 3. A model of the relationship of different types of organisms. All organisms on earth can be organized into a forest of family trees. Organisms on each tree are all offspring of one or a population of ancestors. Lines in between different family trees link characters (phenotypes or genes) that are shared among organisms that do not belong to the same family trees.
Conclusions
The unbridgeable gaps between various biological systems are not consistent with the explanation of random mutation and natural selection through eons of time, because the issue of how gaps can be crossed is an issue not of time but of the abundant and various discontinuities. In the new model of life, organisms are perceived to be modular in nature. Different organisms are linked by a forest of family trees. Different family trees can share similar morphological or genetic features. The identification of such features will be very useful for dissecting signal pathways and molecular circuits controlling the development and/or functions of different biological structures. Allowing organisms to be assigned to different family trees will free molecular phylogeneticists from the burden of having to fit all organisms into one giant tree, and being required in the process to throw away most of the information when performing genomic comparisons, and from having to deal with phylogenetic incongruities or incomplete lineage sorting. The hope is to develop a natural way of categorizing and understanding diverse organisms, using all information encoded in different genomes instead of being forced by the single-tree-of-life model into cherry picking.
References
Axe, D. D. 2004. “Estimating the Prevalence of Protein Sequences Adopting Functional Enzyme Folds.” Journal of Molecular Biology 341 (5): 1295–315. doi:10.1016/j.jmb.2004.06.058.
Clare, J., K. Sreekrishna, and M. Romanos. 1998. “Expression of Tetanus Toxin Fragment C.” In Pichia Protocols: Methods in Molecular Biology, edited by D. R. Higgins and J. M. Cregg, 193–208. Totowa, New Jersey: Humana Press Inc.
Darwin, C. 1859. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London, United Kingdom: John Murray.
Dayrat, B. 2003. “The Roots of Phylogeny: How did Haeckel Build his Trees?” Systematic Biology 52 (4): 515–527.
Eldredge, N., and S. J. Gould. 1972. “Punctuated Equilibria: An Alternative to Phyletic Gradualism.” In Models in Paleobiology, 82–115. San Francisco, California: Freeman Cooper and Co.
Frair, W. 2000. “Baraminology—Classification of Created Organisms.” Creation Research Society Quarterly 37 (2): 82–91.
Frey, E., and H. Tischlinger. 2012. “The Late Jurassic pterosaur Rhamphorhynchus, a Frequent Victim of the Ganoid Fish Aspidorhynchus?” PLoS One 7 (3): e31945. doi:10.1371/journal.pone.0031945.
Gauger, A. K., S. Ebnet, P. F. Fahey, and R. Seelke. 2010. “Reductive Evolution Can Prevent Populations from Taking Simple Adaptive Paths to High Fitness.” BIO-Complexity (2): 1–9.
Gibson, D. G., G. A. Benders, C. Andrews-Pfannkoch, E. A. Denisova, H. Baden-Tillson, J. Zaveri, T. B. Stockwell, et al. 2008. “Complete Chemical Synthesis, Assembly, and Cloning of a Mycoplasma genitalium Genome.” Science 319 (5867): 1215–1220. doi:1151721 [pii] 10.1126/science.1151721.
Gibson, D. G., J. I. Glass, C. Lartigue, V. N. Noskov, R. Y. Chuang, M. A. Algire, G. A. Benders, et al. 2010. “Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome.” Science 329 (5987): 52–56. doi:science.1190719 [pii]10.1126/science.1190719.
Itaya, M., K. Tsuge, M. Koizumi, and K. Fujita. 2005. “Combining Two Genomes in One Cell: Stable Cloning of the Synechocystis PCC6803 Genome in the Bacillus subtilis 168 Genome.” Proceedings of the National Academy of Sciences USA 102 (44): 15971–15976. doi:0503868102 [pii].1073/pnas.0503868102.
Javorsek, D., P. Sturrock, J. Buncher, E. Fischbach, T. Gruenwald, A. Hoft, T. Horan, et al. 2009. “Investigation of Periodic Nuclear Decay Data with Spectral Analysis Techniques.” Intersections of Particle and Nuclear Physics 1182 (1): 292–295.
Javorsek, D., P. A. Sturrock, R. N. Lasenby, A. N. Lasenby, J. B. Buncher, E. Fischbach, J. T. Gruenwald, et al. 2010. “Power Spectrum Analyses of Nuclear Decay Rates.” Astroparticle Physics 34 (3): 173–178. doi:10.1016/j.astropartphys.2010.06.011.
Javorsek, D., M. C. Brewer, J. B. Buncher, E. Fischbach, J. T. Gruenwald, J. Heim, A. W. Hoft, et al. 2012. “Study of Nuclear Decays During a Solar Eclipse: Thule Greenland 2008.” Astrophysics and Space Science 342 (1): 9–13. doi:10.1007/s10509-012-1148-9.
Karas, B. J., C. Tagwerker, I. T. Yonemoto, C. A. Hutchison III, and H. O. Smith. 2012. “Cloning the Acholeplasma laidlawii PG-8A Genome in Saccharomyces cerevisiae as a Yeast Centromeric Plasmid.” ACS Synthetic Biology 1 (1): 22–28. doi:10.1021/sb200013j.
Lartigue, C., A. Blanchard, J. Renaudin, F. Thiaucourt, and P. Sirand-Pugnet. 2003. “Host Specificity of Mollicutes oriC Plasmids: Functional Analysis of Replication Origin.” Nucleic Acids Research 31 (22): 6610–6618.
Lartigue, C., S. Vashee, M. A. Algire, R. Y. Chuang, G. A. Benders, L. Ma, V. N. Noskov, et al. 2009. “Creating Bacterial Strains from Genomes that have been Cloned and Engineered in Yeast.” Science 325 (5948): 1693–1696. doi:10.1126/science.1173759.
Lightner, J. K. 2006. “Identification of Species Within the Sheep-Goat Kind (Tsoan Monobaramin).” Journal of Creation 20 (3): 61–65.
Lightner, J. K. 2007. “Identification of Species Within the Cattle Monobaramin (Kind).” Journal of Creation 21 (1): 119–122.
Lightner, J. K. 2010. “Identification of a Large Sparrow-Finch Monobaramin in Perching Birds (Aves: Passeriformes).” Journal of Creation 24 (3): 117–121.
Lightner, J. K. 2011. “Determining the Ark Kinds.” Answers Research Journal 4: 195–201.
Lightner, J. K. 2012. “Mammalian Ark Kinds.” Answers Research Journal 5: 151–204.
Linnaeus, C. 1758. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, cum Characteribus, Differentiis, Synonymis, Locis. Holmiae, Stockholm: Laurentii Salvii.
Linsell, R. n.d. “The Age of the Universe.” http://www.fizzics.org/Pages/Hubble%2c-doppler%2c-big-bang.aspx. Accessed 10/2.
Linsell, R. 2014. https://www.youtube.com/watch?v=orw4MuprHsY.
Makoff, A. J., M. D. Oxer, M. A. Romanos, N. F. Fairweather, and S. Ballantine. 1989. “Expression of Tetanus Toxin Fragment C in E. coli: High Level Expression by Removing Rare Codons.” Nucleic Acids Research 17 (24): 10191–10202.
Meyer, S. C., M. Ross, P. A. Nelson, and P. Chien. 2003. “The Cambrian Explosion: Biology’s Big Bang.” In Darwinism, Design and Public Education. Edited by J. A. Campbell and S. C. Meyer, 323–402. East Lansing, Michigan: Michigan State University Press.
Meyer, S. C. 2013. Darwin’s Doubt: The Explosive Origin of Animal Life and the Case for Intelligent Design. New York, New York: HarperOne.
Morris, J. D., and F. J. Sherwin. 2013. The Fossil Record: Unearthing Nature’s History of Life. Dallas, Texas: Institute for Creation Research.
Noskov, V. N., B. J. Karas, L. Young, R.-Y. Chuang, D. G. Gibson, Y.-C. Lin, J. Stam, et al. 2012. “Assembly of Large, High G+C bacterial DNA Fragments in Yeast.” ACS Synthetic Biology 1 (7): 267–273. doi:10.1021/sb3000194.
Parker, J., G. Tsagkogeorga, J. A. Cotton, Y. Liu, P. Provero, E. Stupka, and S. J. Rossiter. 2013. “Genome-Wide Signatures of Convergent Evolution in Echolocating Mammals.” Nature 502 (7470): 228–231. doi:10.1038/nature12511.
Patterson, C. 1956. “Age of Meteorites and the Earth.” Geochimica et Cosmochimica Acta 10: 230–237.
Patterson, C., G. Tilton, and M. Inghram. 1955. “Age of the Earth.” Science 121 (3134): 69–75. doi:10.1126/science.121.3134.69.
Puigbò, P., Y. I. Wolf, and E. V. Koonin. 2009. “Search for a ‘Tree of Life’ in the Thicket of the Phylogenetic Forest.” Journal of Biology 8 (6): 59. doi:10.1186/jbiol159.
ReMine, W. J. 1990. “Discontinuity Systematics: A New Methodology of Biosystematics Relevant to the Creation Model.” In Proceedings of the Second International Conference on Creationism. Edited by R. E. Walsh and C. L. Brooks, 207–216. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Robinson, D. A., and D. P. Cavanaugh. 1998. “A Quantitative Approach to Baraminology With Examples from the Catarrhine Primates.” Creation Research Society Quarterly 34 (4): 196–208.
Romanos, M. A., A. J. Makoff, N. F. Fairweather, K. M. Beesley, D. E. Slater, F. B. Rayment, M. M. Payne, and J. J. Clare. 1991. “Expression of Tetanus Toxin Fragment C in Yeast: Gene Synthesis is Required to Eliminate Fortuitous Polyadenylation Sites in AT-rich DNA.” Nucleic Acids Research 19 (7): 1461–1467.
Sandage, A. 1958. “Current Problems in the Extragalactic Distance Scale.” Astrophysical Journal 127 (3): 513–526. doi:10.1086/146483.
Sanders, R. W., and K. P. Wise. 2003. “The Cognitum: A Perception-Dependent Concept Needed in Baraminology.” In Proceedings of the Fifth International Conference on Creationism. Edited by R. L. Ivey, 445–455. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Sorek, R., Y. Zhu, C. J. Creevey, M. P. Francino, P. Bork, and E. M. Rubin. 2007. “Genome-Wide Experimental Determination of Barriers to Horizontal Gene Transfer.” Science 318 (5855): 1449–1452. doi:10.1126/science.1147112.
Steinitz, G., O. Piatibratova, and P. Kotlarsky. 2011. “Possible Effect of Solar Tides on Radon Signals.” Journal of Environmental Radioactivity 102 (8): 749–765. doi:10.1016/j.jenvrad.2011.04.002.
Sverdlov, E. D. 2006. “Is Biological Reductionism Losing Ground? What is Next?” Herald of the Russian Academy of Sciences 76 (4): 339–351. doi:10.1134/S1019331606040058.
Tagwerker, C., C. L. Dupont, B. J. Karas, L. Ma, R. Y. Chuang, G. A. Benders, A. Ramon, et al. 2012. “Sequence Analysis of a Complete 1.66 Mb Prochlorococcus marinus MED4 Genome Cloned in Yeast.” Nucleic Acids Research 40 (20): 10375–10383. doi:10.1093/nar/gks823.
Tan, C. 2015. “Using Taxonomically Restricted Essential Genes to Determine Whether Two Organisms Can Belong to the Same Family Tree.” Answers Research Journal 8: 413–435.
Tan, C., and J. P. Tomkins. 2015a. “Information Processing Differences Between Archaea and Eukarya—Implications for Homologs and the Myth of Eukaryogenesis.” Answers Research Journal 8: 121–141.
Tan, C., and J. P. Tomkins. 2015b. “Information Processing Differences Between Bacteria and Eukarya—Implications for the Myth of Eukaryogenesis.” Answers Research Journal 8: 143–162.
Taylor, S. V., K. U. Walter, P. Kast, and D. Hilvert. 2001. “Searching Sequence Space for Protein Catalysts.” Proceedings of the National Academy of Sciences USA 98 (19): 10596–10601. doi:10.1073/pnas.191159298.
Tomkins, J. P. 2013. “Comprehensive Analysis of Chimpanzee and Human Chromosomes Reveals Average DNA Similarity of 70%.” Answers Research Journal 6: 63–69.
Tregoning, J. S., P. Nixon, H. Kuroda, Z. Svab, S. Clare, F. Bowe, N. Fairweather, et al. 2003. “Expression of Tetanus Toxin Fragment C in Tobacco Chloroplasts.” Nucleic Acids Research 31 (4): 1174–1179.
Vardiman, L., A. A. Snelling, and E. F. Chaffin, eds. 2000. Radioisotopes and the Age of the Earth: A Young-Earth Creationist Research Initiative. El Cajon, California: Institute for Creation Research, and Chino Valley, Arizona: Creation Research Society.
Vardiman, L., A. A. Snelling, and E. F. Chaffin, eds. 2005. Radioisotopes and the Age of the Earth: Results of a Young-Earth Creationist Research Initiative. El Cajon, California: Institute for Creation Research, and Chino Valley, Arizona: Creation Research Society.
Wise, K. P. 1990. “Baraminology: A Young-Earth Creation Biosystematic Method.” In Proceedings of the Second International Conference on Creationism. Edited by C. L. Brooks and R. E. Walsh, 345–358. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Woese, C. R. 2000. “Interpreting the Universal Phylogenetic Tree.” Proceedings of the National Academy of Sciences USA 97 (15): 8392–8396.
Woese, C. R., and G. E. Fox. 1977. “Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms.” Proceedings of the National Academy of Sciences USA 74 (11): 5088–5090.
Wood, T. C. 2006. “The Current Status of Baraminology.” Creation Research Society Quarterly 43 (3): 149–158.
Wood, T. C. 2013. “Mitochondrial DNA Analysis of Three Terrestrial Mammal Baramins (Equidae, Felidae, and Canidae) Implies an Accelerated Mutation Rate Near the Time of the Flood.” In Proceedings of the Seventh International Conference on Creationism. Edited by M. Horstmeyer. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Wood, T. C., K. P. Wise, R. Sanders, and N. Doran. 2003. “A Refined Baramin Concept.” Occasional Papers of the Baraminology Study Group 3: 1–14.
The Age of the Universe
The most popular estimate of the age of the universe is 13.8 billion years. This is based on the value of the Hubble constant (Linsell 2014) (Fig. 4). Fig. 4 is derived from a YouTube presentation by Roger Linsell (https://www.youtube.com/watch?v=orw4MuprHsY). This presentation was published on July 28, 2014 and had been viewed by 4530 people by March 18, 2016. To determine how that number would change, I visited that site a month later (April 19, 2016) and found that number of viewers had increased to 5052. Many others share Linsell’s view on the calculation of the age of the universe, for example, Mike Guidry at University of Tennessee (http://csep10.phys.utk.edu/astr162/lect/cosmology/age.html), Carl Nave at Georgia State University (http://hyperphysics.phy-astr.gsu.edu/hbase/astro/uniage.html), Christopher Palma at Pennsylvania State University (https://www.e-education.psu.edu/astro801/content/l10_p5.html), Oliver Fraser at University of Washington (http://www.astro.washington.edu/courses/labs/clearinghouse/labs/HubbleLaw/hubbles_law_procedure.html), and NASA (National Aeronautics and Space Administration, http://map.gsfc.nasa.gov/universe/uni_age.html). His presentation is chosen because it is detailed and clear.
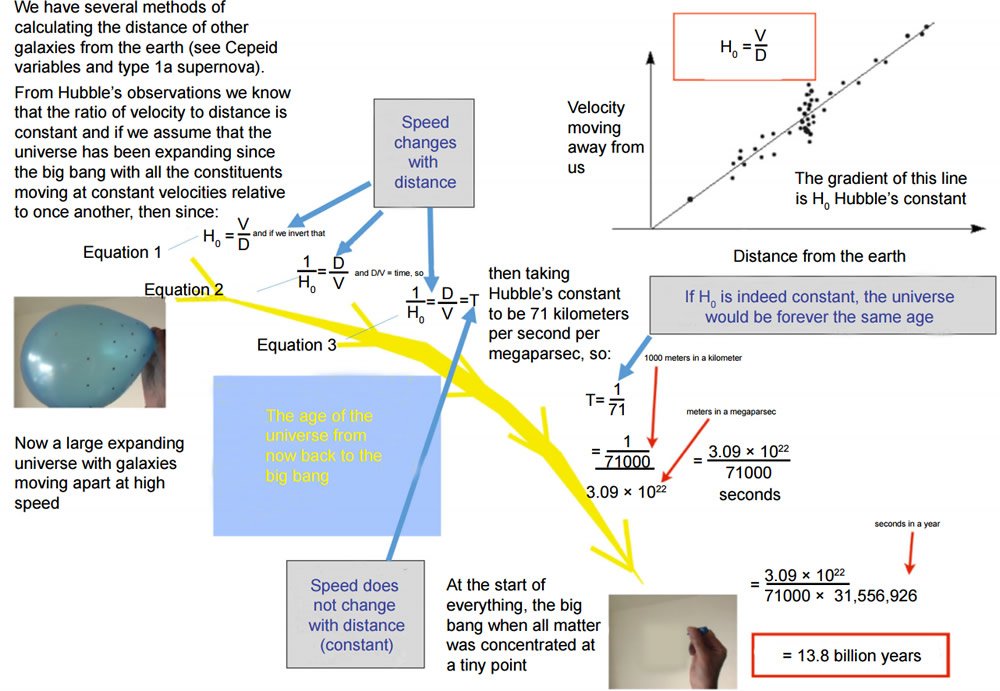
Fig. 4. Estimating the age of the universe using the Hubble’s constant. All items, except the blue arrows and the boxed purple comments, are from (Linsell 2014).
Linsell’s equation 1 is Hubble’s law, which states that (1) objects observed in deep space (extragalactic space, ~10 megaparsecs or more) have a redshift interpretable as relative velocity receding from the earth; and (2) this redshift-measured velocity (V) of various galaxies is approximately proportional to their distance (D) from the earth. H0 is the Hubble constant. Note that one essential assumption here, and in equation 2, which is an inversion of equation 1, is that the observed redshift is caused by the receding of the objects from the earth, instead of other reasons, and our estimates of the distance of the objects are accurate. To estimate the age of the universe, equation 3 is used. The left side of equation 3 is equation 2. For the right side of equation 3 to hold true, a necessary condition is that the object has been moving at a constant speed throughout the whole journey. Another assumption is that the big bang theory is correct and that the current distance of a galaxy from the earth is the distance the galaxy has travelled since the beginning of time. There is a serious problem here regarding distance and time. The theory claims that the distances between the faraway galaxies and the earth we measured are current. And yet the same theory also tells us that we are looking back in time, billions years of time. Therefore, those objects may not exist anymore. If they do exist, it will take additional billions of years for us to see their current status, according to the same theory). Note that according to the big bang theory, all things were together at a single point at the beginning, thus, their relative speed was zero at that time. However, equations 1 and 2 require things to move with different speed, while equation 3 requires things to move at constant speed. In addition, even if equation 1 is correct, that does not mean equation 3 is correct; the movement of the objects does not even need to be continuous, not to say at constant speed. Indeed, scientists used to believe that the expansion of the universe was slowing down due to gravity and now think that the expansion is speeding up because of dark energy. Furthermore, if the Hubble constant were indeed constant and equation 4 were true, the universe would stay forever as 13.8 billion years old because the inverse of a constant is a constant, so it will not change with time. A universe with an unchangeable age is absurd! (Well, the number of H0 has been chosen to get 13.8 billion years for the age of the universe. The current accepted value is a little less than this, so another constant is normally introduced to make the age and the Hubble time agree. Nonetheless, the introduction of another constant does not affect the argument.)
My point is that current scientific knowledge or methods actually do not allow us to know the age of the universe. All age estimations are plagued with assumptions. Just as Allan Sandage in his Helen Warner Prize lecture of the American Astronomical Society said “The numerical value of H [H0 in Fig. 4] is important because it tests the time-scale criteria in various world models. By itself, H does not give the time since the ‘beginning’ of the expansion. Knowledge of the world model must also be available” (Sandage 1958). Thus, to estimate the age of the universe, a scientist has to use H0 together with a physical theory and assumptions.
The Age of the Earth
Radioactive Dating
The most commonly advocated age of the earth is 4.56 billion years, which is based on Pb-Pb isochron radioactive dating of three stony and two iron meteorites, plus an oceanic sediment sample whose lead was assumed to have been generated via radioactive decay of uranium over earth time by Patterson and his colleagues (Patterson 1956; Patterson, Tilton, and Inghram 1955). Two main assumptions for the dating are that “when the earth was formed, it contained lead with an isotopic composition the same as that found in iron meteorites, and that the ratio of lead to uranium has not changed, except for radioactive decay in the surface of the earth, since the surface was formed” (Patterson, Tilton, and Inghram 1955, 69). That is, (1) we know the initial conditions, and (2) we are dealing with a closed system where no parental or daughter elements were gained or lost by any means except radioactive decay since the birth of the earth. What is the basis for such an assumption? Patterson and his colleagues said bluntly: “There is no evidence, therefore, that compels us to assume that when the earth was formed it contained lead with the same isotopic composition as that in iron meteorites, although such an assumption is necessary for the age calculations” (Patterson, Tilton and Inghram 1955, 70). In addition, several assumptions were made about the meteorites: “they were formed at the same time; they existed as isolated and closed systems; they originally contained lead of the same isotopic composition; they contain uranium which has the same isotopic composition as that in the earth” (Patterson 1956, 230–231). Therefore, it was assumed, out of necessity for the estimation, that both the earth and the meteorites have to be closed systems.
The assumption of the initial conditions will forever remain as guesswork because nobody can go back into history to investigate. In contrast, the second assumption, that we are dealing with closed systems, can be tested by observation. In fact, Patterson and his colleagues found that radiogenic lead varies greatly in the crust of the earth (Patterson, Tilton, and Inghram 1955): radiogenic 206Pb is 40% in excess and 208Pb 80% in excess in basalt, while they are 50% deficient and 80% deficient, respectively, in granite. It is a “good” and “extensive mixing” of the materials that generated the oceanic sediment lead that was used to deduce the age of the earth. That is, the earth is an open system.
Furthermore, a casual examination of the radioactive decay chains reveals that the assumption of a closed system contradicts with the chemical natures of the decay intermediates. Lead-206 (232Pb) is the final product of the multiple decay steps of uranium-238 (238U, Fig. 5A), while two other isotopes of lead, 207Pb and 208Pb, are the final products of the multiple decay steps of 235U (Fig. 5B) and thorium-232 (232Th, Fig. 5C), respectively. Note that a common intermediate element of these decay series is radon, a noble gas. Therefore, it normally does not bind chemically to other elements, and, as any other gas, it does not stay where it is. This is why before people decide to buy a house they want the house to be inspected to see whether there is too much radon in the house to avoid inhalation of the dangerous radioactive radon gas. Another intermediate decay product of 238U is mercury (Hg, Fig. 5A), which is a liquid at room temperature and also evaporates at room temperature. Breathing air contaminated with mercury vapor can cause mercury poisoning. Therefore, neither the uranium nor the thorium decay chains can be closed systems due to the innate chemical natures of the decay intermediates.
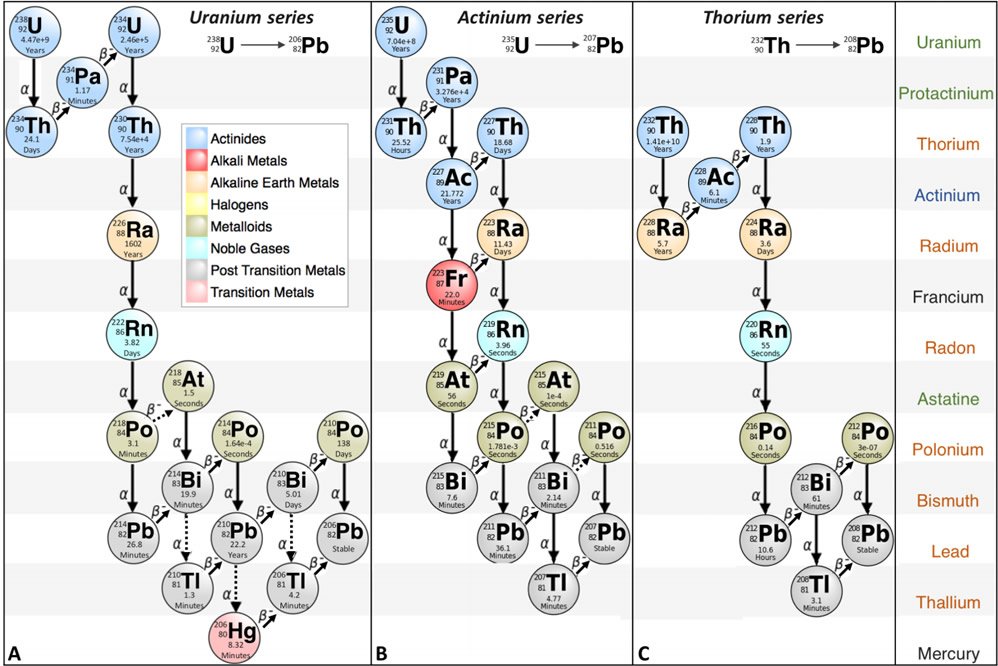
Fig. 5. Decay series of (A) uranium-238 (238U), (B) uranium-235 (235U), and (C) thorium-232 (232Th). Full names of elements shared by all three decay series are in coral, by the two uranium series in green, by 235U and 232Th in blue, and unique to 238U or 235U in black. The images are modified from the decay chains at https://en.wikipedia.org/wiki/Decay_chain.
Other possible processes which would make the meteorites or the earth rocks which are used to estimate the age of the earth open systems include earthquakes, volcanic eruptions, weathering, and processes occurring in the sun or other celestial bodies (as discussed below).
Another unstated assumption is that the radioactive decay rates are constant. This assumption has recently been found to be false. In fact, the decay rates of two of the intermediate decay products of the 238U, 235U, and 232Th decay series, radium and radon, have been observed to vary with activities of the Sun, though how this occurs is still not known (Javorsek et al. 2012; Javorsek et al. 2009, 2010; Steinitz, Piatibratova, and Kotlarsky 2011). Furthermore, it seems that the decay rates of radioactive materials were much faster in the past (Vardiman, Snelling and Chaffin 2000, 2005).
Therefore, the estimate of the age of the earth by radioactive dating is also based on assumptions; one of them, the initial conditions, is untestable, and the other two cannot be reconciled with the innate chemical natures of the decay intermediates and recent experimental observations. Thus, both the estimate of the age of the earth based on radioactive decay and the estimation of the age of the universe using the Hubble constant are theory/assumptions-based and are questionable.
One may argue that the universe and the earth are at least as old as shown by the fossil record. Therefore, we will next look at the fossil record and see what this record teaches.
The Fossil Record
I used to think that one has to dig hard to find any fossils. But I discovered that fossils are everywhere. In fact, I am surrounded by fossils every day. I live on a campus, the University of Missouri (MU), which is full of fossils, in a city, Columbia Missouri, which is full of fossils. For example, on the MU campus there are many stone buildings. All of the stones, natural stones, not the artificial ones, which I have checked have crinoid fossils in them. Crinoid fossils are the state fossil of Missouri. At one time believed to be extinct 300 million years ago, crinoids have been found living, and there are more than 600 species of them (https://www.youtube.com/watch?v=IFWeqDcAYGk, a live video on “Feather Stars and Sea Lilies” by Monterey Bay Aquarium, see also https://en.wikipedia.org/wiki/Crinoid). Photos of a few coral, bivalve, brachiopod, and crinoid fossils I have seen in the stones of MU are shown in Fig. 6.
What the fossil record shows are the following:
- The sudden appearance of completely formed animals and plants
- Extinction of species
- Stasis of species (The norm for a species is stability)
- Catastrophic burial of live organisms
The catastrophic burial of live organisms can be vividly seen in the fossils of a dinosaur Sinosauropteryx prima with a fish, of an ichthyosaur giving birth, of a fish eating another fish (Morris and Sherwin 2013), and of a pterosaur Rhamphorhynchus with a ganoid fish Aspidorhynchus (Frey and Tischlinger 2012). The Rhamphorhynchus pterosaur seems to have been caught alive, with one fish still in its throat, by the ganoid fish Aspidorhynchus.
As is the case with the estimate of the age of the universe according to the Hubble constant or the age of the earth according to the radioactive decay of uranium, fossils themselves do not tell us the age of the earth. Any age association with the fossils is dependent upon the presuppositions of the investigators.
As Eldredge and Gould stated, “We do not encounter facts as data discovered objectively. All observation is colored by theory and expectation . . . Theory does not develop as a simple and logical extension of observation; it does not arise merely from the patient accumulation of facts. Rather, we observe in order to test hypotheses and examine their consequences . . . In paleontology, even the most ‘objective’ undertaking, the ‘pure’ description of fossils, is all the more affected by theory because that theory is unacknowledged. We describe part by part and are led, subtly but surely, to the view that complexity is irreducible . . . The inductivist view forces us into a vicious circle. A theory often compels us to see the world in its light and support. Yet, we think we see objectively and therefore interpret each new datum as an independent confirmation of our theory. Although our theory may be wrong, we cannot confute it” (Eldredge and Gould 1972, 85–86, emphasis added). When mentioning the impact of textbooks on the thoughts of new professionals, they quoted comments of Kuhn—The “normal science” that they inculcate is “a strenuous and devoted attempt to force nature into the conceptual boxes supplied by professional education” (Eldredge and Gould 1972, 91), and later added “That these older texts hold so strongly to phyletic gradualism should surprise no one; harder to understand is the fact that virtually all modern texts repeat the same arguments even though their warrant had disappeared” (Eldredge and Gould 1972, 92).
Finally, not only has the age of the universe or of the earth not been established by scientific observation, as we are so often told, but it cannot be. This is determined by the nature of history: history goes in only one direction and cannot be repeated. Instead, I have argued here that the main issue at hand, regarding the origin of life and the origin of the diversity of life, is not time but discontinuity between different types of life forms, such as the unbridgeable gaps between prokaryotes and eukaryotes.