The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
No known molecule, including DNA, RNA, and proteins, can replicate itself; dozens of specific proteins are required to replicate a small bacterial genome. DNA, however long it is and however many genes it can encode, is nothing without the molecular machineries to decode (that is, to transcribe and translate) its encoded genes. Amazingly, each of the three domains of life (bacteria, archaea, and eukaryotes) has its own unique way of replicating its genome, defining whether a piece of DNA is a gene or not, whether an RNA is protein-coding or not, and where transcription or translation should start and end. This creates unbridgeable gaps in between bacteria, archaea, and eukaryotes and, thus, challenges the popular belief that life came from non-life naturally and that all organisms are connected via a big evolutionary tree of life.
Keywords: abiogenesis, origin of life, information coding and decoding, DNA replication, gene transcription, gene translation
Introduction
As I mentioned in #1 and #2 of this series on Facts Cannot be Ignored When Considering the Origin of Life (Tan 2022a, 2022b), an astonishing discovery of molecular biology is organism-specific biological information coding and decoding systems. This was what started this author to inquire the origin of eukaryotes and the origin of life sixteen years ago. Biological information coding and decoding refers to DNA replication (the process that two copies of genomic DNA are made from one copy), gene transcription (making RNA from DNA templates), and gene translation (making proteins whose amino acid sequences are determined by the nucleotide sequences of RNA). It is impossible to fully address this discovery in one short article. This essay is merely a foretaste of what I hope to detail in the future. Specifically, a brief comparison of bacterial and eukaryotic DNA replication initiation, transcription initiation, and translation initiation will be presented to illustrate how the same vital tasks of replicating genomic DNA and transcribing and translating genes are implemented differently in different domains of life using proteins that are mostly unrelated in amino acids sequence.
A comparison of bacterial and eukaryotic DNA replication initiation
DNA replication is indispensable for the survival and reproduction of every organism, without exception. To compare and contrast bacterial and eukaryotic DNA replication initiation, I determined the distribution of homologs of proteins involved in the initiation of bacterial and eukaryotic DNA replication. A homolog of a protein of interest is commonly regarded as a protein shared a common ancestorial gene with the protein of interest, however here it is defined as a protein with some sequence similarity with the protein of interest, regardless its origin.
The results are shown in Fig. 1. Each row represents one specific gene, with its total homolog number next to the name of the gene. Each column represents a group of organisms with the numbers of species analyzed in that group listed underneath the name of that group. The number at the intersection of a row and a column is the number of species in the group of that column that contain homologs to the gene of that row. The percentage of species with homologs for a specific gene in a group are color-coded (red: 0, none species in the group contains a homolog for that gene; green: 100%, every species in the group contains a homolog for that gene; different shades of mixed red and green: frequency between 0 and 100%, the higher the frequency, the greener the color). Orphan genes, genes with no homologs in all the organisms (other than the reference organism itself) compared, are marked with red stars. Nearly orphan genes, genes with homologs in 1–5 species (other than the reference organism) of all species compared, are marked with blue stars.
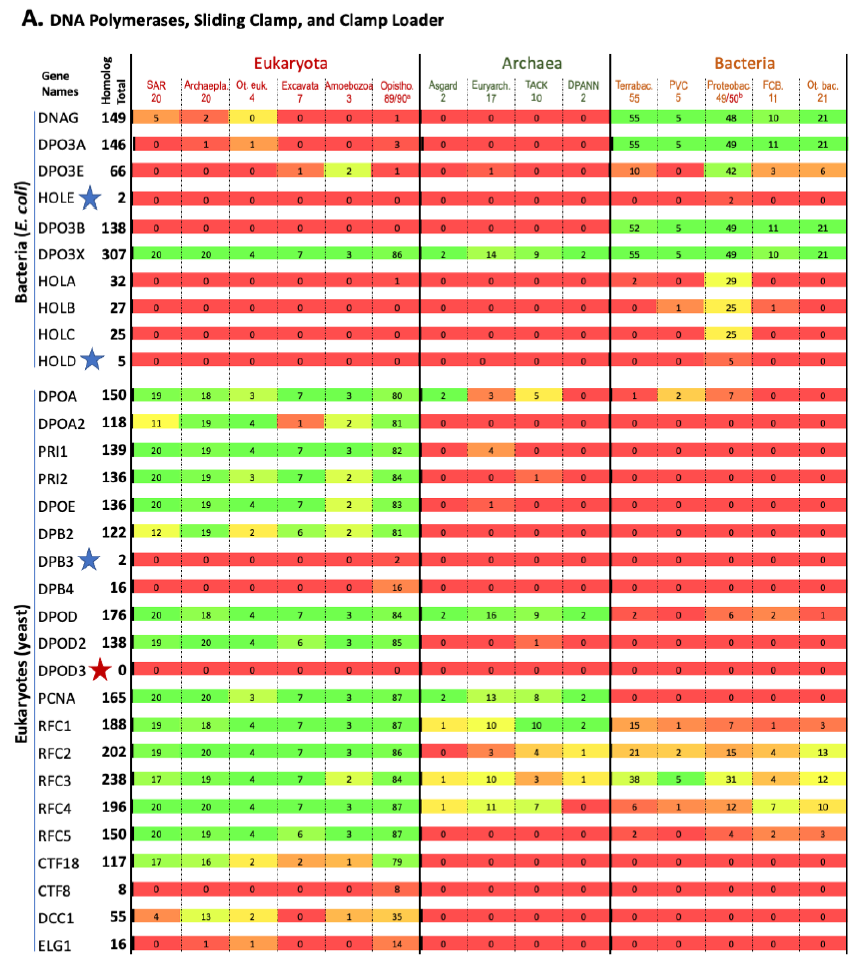
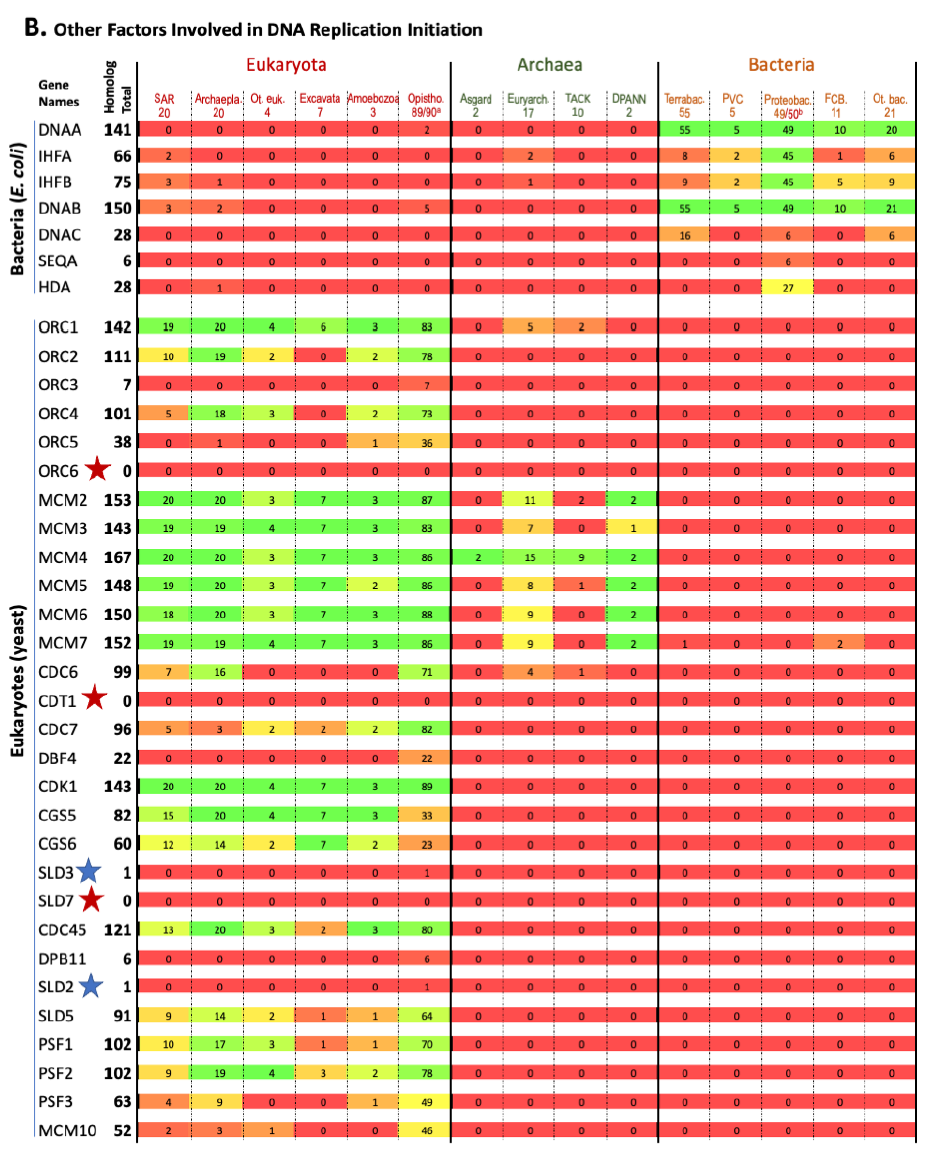
Fig. 1. Homolog distribution of genes involved in bacterial and eukaryotic DNA replication initiation.
(A page 50) Distribution of homologs of DNA polymerases, sliding clamps, and clamp loaders.
(B page 51) Distribution of homologs of other factors involved in DNA replication initiation.
Method: To compare the DNA replication machineries of bacteria and eukaryotes, homologs of E. coli and Saccharomyces cerevisiae proteins involved in DNA replication are searched against the 317 (144 eukaryotes, 142 bacteria, and 31 archaea) three domain model organisms database and either the bacteria-only (3863 bacterial species ) or eukaryotes-only (711 eukaryotes) databases in the OrthoInspector website (Nevers et al. 2019).
Note: a. The budding yeast S. cerevisiae is one of the 90 Opistho species in the database. Thus, only 89 of them should be counted when searching homologs of a yeast gene. b. E. coli is one of the 50 Proteobac species in the database. Thus, only 49 of them should be counted when searching homologs of an E. coli gene.
Abbreviations of organism groups (in the order of appearance): Eukaryotes: SAR: Stramenopiles, Alveolata, Rhizaria; Archaepla: Archaeplastida; Ot. euk: Other Eukaryota; Opistho: Opisthokonta. Archaea: Asgard: Asgard group (including Lokiarchaeota and Thorarchaeota); Euryarch: Euryarchaeota; TACK: TACK group (including Thaumarchaeota, Bathyarchaeota, Crenarchaeota, and Korarchaeota); DPANN: DPANN group (including Nanoarchaeota). Bacteria: Terrabac: Terrabacteria; PVC: a superphylum of bacteria including Planctomycetes, Verrucomicrobia, and Chlamydiae; Proteobac: Proteobacteria; FCB: a superphylum of bacteria including Fibrobacteres, Chlorobi, and Bacteroidetes; Ot. bac: Other Bacteria.
Fig. 1 shows that a species that is colored green on one side (right or left with bacteria on the right, eukaryotes on the left, and archaea in the middle) is generally red on the other side. This demonstrates that most proteins used by bacteria to replicate their genomes do not have homologs in eukaryotes, and vice versa. Thus, bacteria use mostly bacteria-unique genes to replicate their genomes, and eukaryotes use mostly eukaryotes-unique genes to replicate their genomes.
This is evidence that the same task is implemented differently by the three fundamental cell types.
This is not what one would expect if bacteria and eukaryotes had shared a common ancestor since DNA replication is essential for the survival and reproduction of each and every known organism. In other words, these bacteria or eukaryotes-unique genes that are needed for DNA replication of their specific domains of life challenge the belief that eukaryotes evolved from bacteria, and the belief that all life forms are connected via an evolutionary tree of life.
A Comparison of Bacterial and Eukaryotic Gene Transcription Initiation
Like DNA replication, gene transcription is indispensable for the survival and reproduction of every organism. Bacteria use one type of RNA polymerase, an enzyme that synthesis RNA using DNA as a template, that is made of five different proteins to transcribe all genes encoded in their genomes. In contrast, eukaryotes use at least three RNA polymerases made of 12–17 different proteins to transcribe their genes, each polymerase responsible for a specific set of genes (fig. 2).
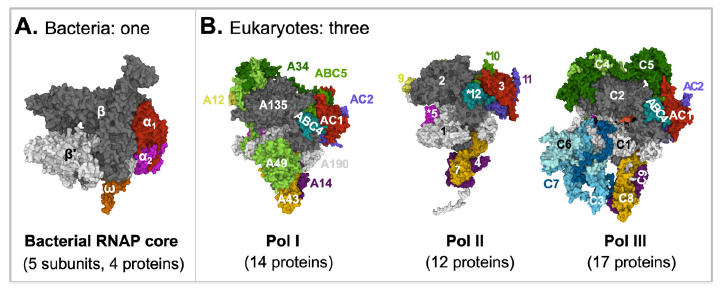
Fig. 2. A comparison of bacterial and eukaryotic RNA polymerase cores. Proteins with sequence similarities are represented with the same colors.
A: Bacterial RNA polymerase core. E. coli RNA polymerase core is made of four proteins: α (two copies), β, β’, and ω. The structure was extracted from (PDB: 6B6H) (Liu et al. 2017).
B: Eukaryotic RNA polymerases. RNA Polymerase I is made of 14 proteins: A190, A135, A49, A43, A34, A14, A12, ABC1-5, AC1, and AC2. RNA Polymerase II is made of 12 proteins: Rpbs 1-12. RNA Polymerase III is made of 17 proteins: ABC1-5, AC1, AC2, and C1-10. The structures are for yeast S. cerevisiae Pol I (PDB: 6RWE) (Sadian et al. 2019), human Pol II (PDB: 7ENC) (Chen, Yin et al. 2021), and human Pol III (PDB: 7AE1) (Girbig et al. 2021).
Figure credits: Images were created at RCSB PDB with Mol* (JavaScript) (Sehnal et al. 2018).
Strikingly, the bacterial RNA polymerase core needs to form a protein complex with one other protein, while the eukaryotic RNA polymerases, which are made of two to three times more proteins, needs to form a complex with multiple general transcription factors to locate gene promoters (fig. 3). Two of the Pol II transcription factors, TFIID and THIIH, are made of 14 and 10 different proteins, respectively. TFIIB is the only Pol II general transcription factor that is made of a single protein.
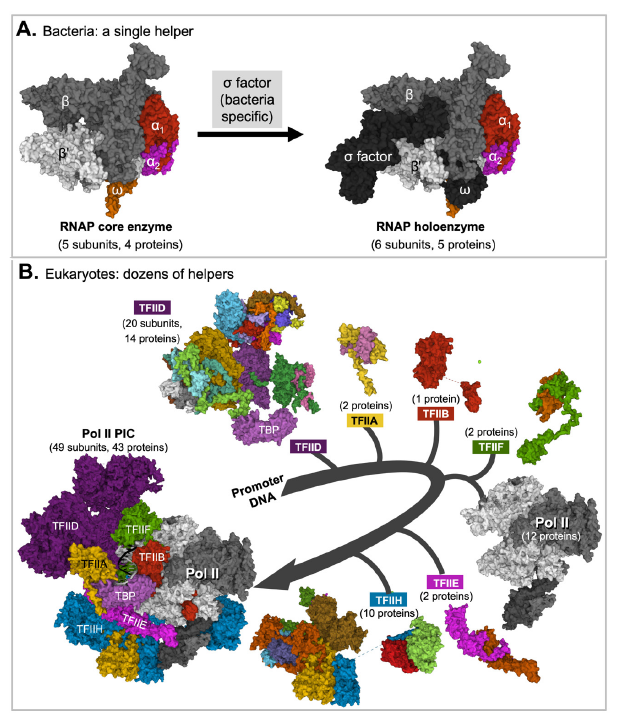
Fig. 3. Eukaryotic RNA polymerases need help from many more proteins than bacterial RNA polymerase core does.
A. A bacterial RNA polymerase core needs help from one protein to initiate transcription. Shown in the figure is the core (left) and holo-enzyme (right) of E. coli RNA polymerase.
B. Eukaryotic RNA Pol II needs help from six general transcription factors made of 31 unique proteins. Note that Pol II and Pol II transcription factor (TFII) A, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH need to be assembled in a sequential manner to form a functional preinitiation complex (PIC) made of 49 subunits and 43 different proteins. The subunits of each transcription factor are represented with different colors when shown in the transcription factor alone but as a single color in the preinitiation complex (PIC), except with TFIID in which the color of its TATA box binding protein (TBP) subunit is unchanged. The structures of individual components shown are those in the assembled complex and are often different from their structures in their unassembled forms. Figure credits: Images were created at RCSB PDB with Mol* (JavaScript) (Sehnal et al. 2018) from (PDB: 6B6H) (Liu et al. 2017) (Panel A) and (PDB: 7EGB) (Chen, Qi et al. 2021) (Panel B).
To ensure that each gene in a eukaryotic cell is expressed when needed, and only when needed, in the cells needed, at the levels needed, gene-specific transcription factors that bind DNA motifs in gene enhancers are needed. However, most gene-specific transcription factors are unable to directly bind, and thus regulate, the basal transcription machinery made of the RNA polymerase and the general transcription factors. This problem is solved in eukaryotic cells using a protein complex called mediator. The mediator does not bind DNA, but it interacts with the transcription factors that can bind DNA. In addition, it interacts, directly, with the basal transcription machinery.
The mediator alone is composed of more than twenty different proteins (the exact number of proteins varies in different organisms). The structures of Pol II pre-initiation complex (PIC) with mediator (except its kinase domain) from human and mouse have been solved (Chen, Yin et al. 2021; Zhao et al. 2021). The structure of the human complex, which contains 68 unique proteins, is shown in (fig. 4). With the inclusion of the mediator and the general transcription factors in the transcription machinery, Pol II itself appears so small and insignificant.
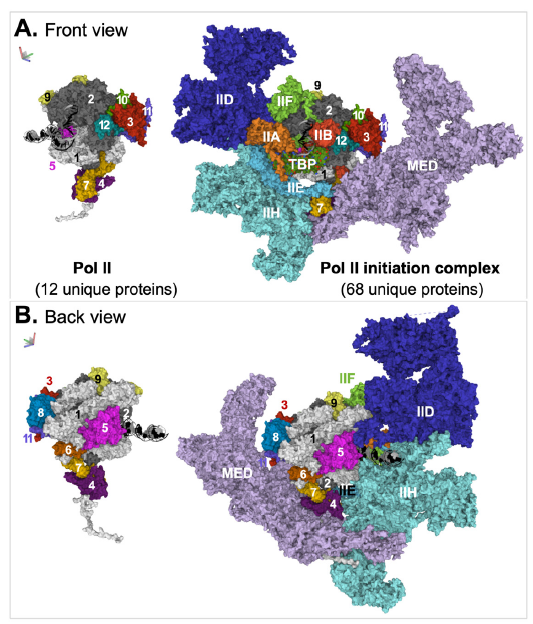
Fig. 4. Structure of the mediator-containing Pol II transcription preinitiation complex. Two views, front (A) and back (B), of the complex are shown. The relative positions of the two views, which are about 180° apart, are indicated by an image of a three-dimensional coordinate on the top left corner of the corresponding panel. Coloring scheme: DNA are shown in cartoon model, with the template strand in black and the non-template strand in gray. TATA box binding protein (TBP) is shown in space-fill model with different colors for different atoms. All other molecules are represented with surface model. The subunits of Pol II are individually colored and numbered. Each transcription factor, except TFIID, is colored with a single color. All subunits of TFIID, except TBP, are colored with one color. The images were generated from PDB: 7ENC (Xizi Chen, Yin et al. 2021) via Mol* (Sehnal et al. 2018). Abbreviations: IIA, IIB, IID, IIE, IIF, IIH: Pol II transcription factor A, B, D, E, F, H; MED: mediator.
In short, bacteria and eukaryotes genes are defined and recognized differently. This is like Chinese and English, they use totally different alphabets, words, and grammars and need to be read differently.
Again, same task, but entirely different implementations.
This creates another challenge to the idea that life came from non-life, that complicated life evolved from simpler ones, and that all life forms are connected via an evolutionary tree of life.
A Comparison of Bacterial and Eukaryotic Gene Translation Initiation
Like DNA replication and gene transcription, gene translation is indispensable for the survival and reproduction of every organism. Gene translation occurs in ribosomes in all organisms. Bacterial ribosomes are made of fifty or so ribosomal proteins and three ribosomal RNAs, while eukaryotic ribosomes are made of about 80 ribosomal proteins and four or more ribosomal RNAs. Some of the bacterial ribosomal proteins are similar in amino acid sequence to eukaryotic ribosomal proteins and some are unique to bacteria.
Strikingly, the three domains of life—bacteria, archaea, and eukaryotes—each has its own peculiar way of determining whether a piece of RNA is protein-coding and, if it is protein-coding, where to start and to stop translation. Fig. 5 provides a comparison of bacterial and eukaryotic translation initiation.
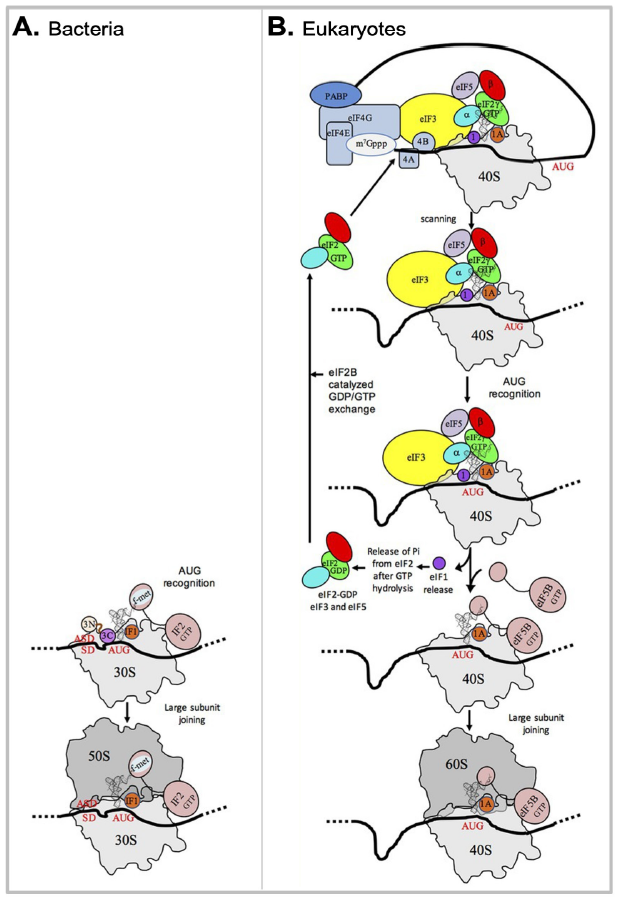
Fig. 5. A comparison of bacterial and eukaryotic gene translation initiation.
(A) Bacterial gene translation initiation. Bacterial 30S subunit recruits the messenger RNA (mRNA), often due to the base pairing between a Shine-Dalgarno sequence (SD) with an anti-SD (ASD) sequence at the 3’-end of 16S rRNA. Three initiation factors, IF1, IF2, and IF3 favor the recruitment of the initiator tRNA and its pairing with the start codon. The formyl-methionyl moiety of the initiator tRNA is important for recognition by IF2. After start codon recognition, IF3 is released and the large ribosomal subunit is recruited with the help of IF2.
(B) Eukaryotic gene translation initiation. Unlike bacteria, eukaryotes do not use the SD anti-SD strategy. In canonical eukaryotic translation, a pre-initiation complex, containing the small ribosomal subunit, the methionylated initiator tRNA, and initiation factors, forms at the 5’-capped end of the mRNA. The complex then scans the mRNA until a start codon in a suitable environment is found. Base-pairing of the tRNA anticodon with the AUG start codon triggers eIF1 release followed by the release of a phosphate group resulting from GTP hydrolysis by eIF2. In turn, eIF2, eIF3, and eIF5 are released; eIF5B-GTP is recruited and favors joining with the large ribosomal subunit. The complex formed by eIF4E + eIF4G + eIF4A is known as eIF4F. eIF3, composed of 6 (yeast) to 13 (mammals) subunits is represented as a yellow oval. Figure credits: modified from Fig. 1 of (Schmitt et al. 2020)(CC BY 4.0).
Note that we only need three fingers to fully count the bacterial translation initiation factors (IFs) (fig. 6). To number the eukaryotic translation initiation factors (eIFs), we will need a six-fingered hand. Furthermore, the fingers need to have branches because we need to count the letters A, B, C, etc. at the same time. Note that the initiation factors whose names containing the same number but different letters are unrelated in protein composition or function, and that each bacterial initiation factor is made of one single protein, while many eukaryotic initiation factors are made of multiple subunits. For example, eukaryotic initiation factor 3, which is represented with an unbranched finger, is made of 6 to 13 different proteins (the number varies depending on the organisms).
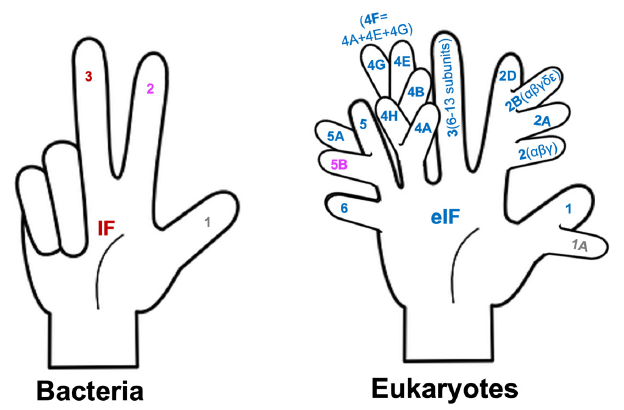
Fig. 6. Counting bacterial and eukaryotic translation initiation factors. Bacterial IF 2 and eukaryotic eIF5B share some sequence similarities, and, thus, are shown with the same color. The same is true for bacterial IF1 and eukaryotic eIF1A.
As DNA replication and gene transcription, the number of proteins involved is not the key issue. The key issue is the identities of the proteins. Both bacteria and eukaryotes require their organism-specific proteins to translate their genes.
Once more, same task, but different implementations.
This creates yet another challenge to the idea that life came from non-life, that complicated life evolved from simpler ones, and that all life forms are connected via an evolutionary tree of life.
A Comparison of Archaeal and Eukaryotic Gene Transcription
It has often been said that archaea can function as intermediates between bacteria and eukaryotes (for example, (Gribaldo et al. 2010; Koonin 2015; Rochette, Brochier-Armanet, and Gouy 2014; Vesteg and Krajcovic 2011; Williams et al. 2013). However, whether eukaryotes are more similar to archaea or more similar to bacteria depend on what genes are compared. For instance, the eukaryotic information processing system is more similar to that of archaea than to that of bacteria, but the eukaryotic metabolic system is more similar to that of bacteria than to that of archaea (Tan and Tomkins 2015a; Tan 2017).
I examined the archaeal and eukaryotic information processing systems and found that they are distinct and unexchangeable (Tan 2017; Tan and Tomkins 2015a). For example, the largest subunit, known as Rpb1, of eukaryotic RNA polymerase II, has homologs in archaea (fig. 7). However, the eukaryotic protein has a unique C-terminal tail (fig. 7, red box) that has no archaeal counterpart. This C-terminal tail is necessary for eukaryotic gene transcription initiation, elongation, and termination. In other words, without the eukaryote-specific C-terminal tail, eukaryotic life would be impossible since its protein-coding genes would not be transcribed, and, consequently, its proteins could not be generated and its genomic DNA whose replication requires proteins could not be replicated.
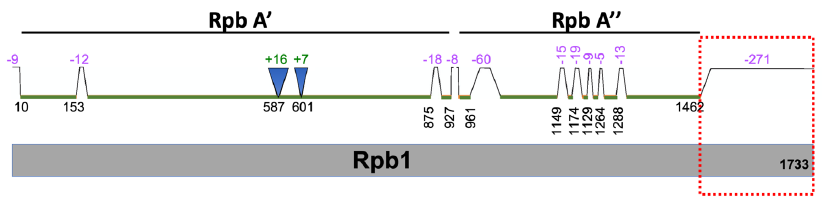
Fig. 7. A comparison of S. cerevisiae Rpb1 (gray, bottom) with its archaeal homologs P. furiosus Rpb A’ and A” (green, top). Note that Rpb A’ is homologous to one part (N-terminal) of Rpb1 and Rpb A” is homologous to another part of Rpb1. Segment locations are based on amino acid positions of Rpb1 protein. Segments present in Rpb1 but not in Rpb A’ and A” are indicated with trapezoids and negative numbers, while those segments absent in Rpb1 but present in Rpb A’ and A” are indicated with inverted triangles and positive numbers. The values of the numbers represent the numbers of amino acid missed and correlate with the sizes of the triangles and trapezoids. The C-terminal tail of Rpb1 that is missing in P. furiosus RNA polymerase is highlighted with a red dotted box.
Organism-Specific Cryptographic Keys
To borrow the language of cryptography, the inheritable genetic information that determines life or death of all living beings is encrypted. Furthermore, the information is encrypted in different ways in different organisms, one way for bacteria, another way for archaea, and yet another way for eukaryotes. These organisms have to use their own unique cryptographic keys to decipher their genomes. These organisms differ in whether they (not we human intellectuals!) regard a segment of DNA as a gene or not, whether a gene is protein-coding or non-protein-coding, where a transcription should start and end, and where a translation should start and end. Their cryptographic keys are their own RNAs and proteins present in their own cells and those that they can make themselves using their own molecular machineries.
For example, the same RNA transcript may encode totally unrelated proteins by a bacterial cell that uses Shine-Dalgarno sequence to identify its translation starting site and a eukaryotic cell that uses a scan mechanism to do that even if it does encode a protein (fig. 8).
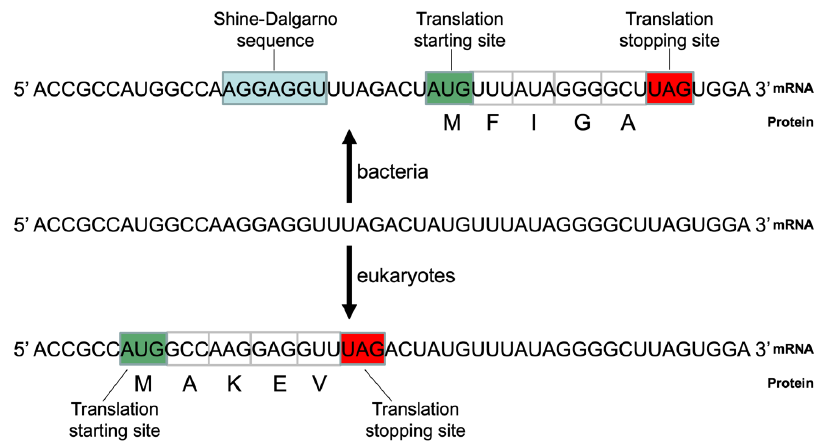
Fig. 8. The same RNA may end up with two different proteins in bacteria and eukaryotes. Blue box: Shine-Dalgarno sequence (SD); green box: translation initiation site; red box: translation stopping site. Top: The hypothetical mRNA would be used to code for a protein with amino acids MFIGA, based on the mechanism of translation of bacteria like E. coli. The SD is important for translation initiation in bacteria. It hybridizes to an anti-SD sequence, which is reverse and complementary to the SD, in the 16S rRNA of the bacterial ribosomes. Bacteria use the AUG that is a few nucleotides downstream of the SD as the translation initiation site. Bottom: The same hypothetical mRNA would be used to code for a protein with amino acids MAKEV, based on the mechanism of translation of eukaryotes like yeast. Eukaryotes normally use the first AUG from the 5’ end of an mRNA as the translation starting site.
Using an analogy to cryptography, the encrypted string of letters “NOT TO BE NOT” can mean quite different, even opposite, things depending on which cryptographic key is used (fig. 9)
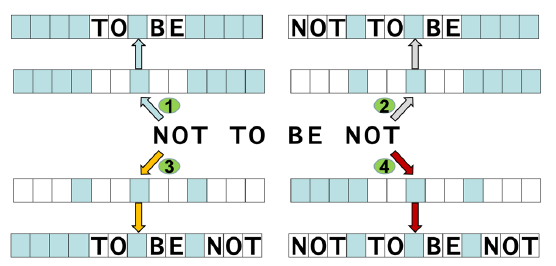
Fig. 9. Decrypting encrypted messages. The same string of letters “NOT TO BE NOT” can mean “TO BE”, “NOT TO BE”, “TO BE NOT”, or “NOT TO BE NOT”, depending on the cryptographic keys of the interpreters. The cryptographic keys are represented with rows of blue and white boxes in which only white boxes allow a letter to be seen.
Mutation and Natural Selection
Could accumulated mutation and natural selection account for the vast number of organism-specific genes necessary for the survival and reproduction of organisms, especially those for their DNA replication, transcription, and translation?
With this question in mind, I have investigated what have been discovered, experimentally, about mutation and natural selection and formation of new genes. The answer for the question is a clear “NO”. Some results of my early investigations were reported in (Tan 2015).
A Logical Conclusion
A logical conclusion from analyses of the genes involved in biological information coding and decoding and of the genes necessary for organism survival and reproduction is that not all organisms can be linked via a big evolutionary tree of life. Organisms on earth are better represented as a forest of trees of life, as I proposed a few years ago (Tan 2016). What many believe and teach about the origin of life and the origin of biodiversity does not agree with what the genes are showing us.
Unfortunately, this conclusion is not one that many would feel comfortable with.
An Experimental Demonstration
The organism-specificity and non-interchangeability, at least at the domain level of life, of biological information coding and decoding systems (Tan and Tomkins 2015a, 2015b), the necessity of many organism-specific essential genes (Tan 2015), and the inability of mutation and natural selection to generate a single essential gene are demonstrated by molecular cloning experiments, especially that of Craig Venter in his creating the “the first self-replicating species we’ve had on the planet whose parent is a computer” (Gibson et al. 2010; Tan 2016; Venter 2010), though whether the parent of the cell they synthesized is a computer is controversial (Matuscak and Tan 2016) and it was not Venter’s intention to provide such a demonstration.
Briefly, Craig Venter and colleagues synthesized the entire one-megabase (Mb) genome of Mycoplasma mycoides in yeast, a eukaryotic cell (Gibson et al. 2010) (fig. 10). However, the yeast cells could not create M. mycoides cells using the cloned bacterial genome. The genes encoded in the cloned genome need to be transcribed and translated using the molecular machines from Mycoplasma capricolum, a cell that shares more than 99% identity for the 79 core proteins involved in gene translation, as well as their ribosomal DNA, with the genome donor M. mycoides (Labroussaa et al. 2016).
Fig. 10. Basic steps involved in Venter’s generation of a synthetic bacterial cell.
The inability of a yeast cell to decode the bacterial M. mycoides genetic code is a consequence of the domain-specific information processing systems, including DNA replication, transcription, and translation (Tan and Tomkins 2015a; 2015b). As mentioned earlier, what is striking is not so much that the number of proteins involved are different (as important as that is) but that the identities of these proteins are different. The proteins used for bacterial DNA replication, transcription, and translation are mostly bacteria specific; they do not have known homologs in eukaryotes. Likewise, the proteins used for eukaryotic DNA replication, transcription, and translation are mostly eukaryotes specific; they do not have known homologs in bacteria.
To overcome the barrier between bacterial and eukaryotic DNA replication machinery, a yeast origin of replication had to be artificially incorporated into the bacterial genome before the bacterial genome could be cloned in yeast (fig. 10, the leftmost arrow on the top). Furthermore, at each step in which the cloned bacterial DNA needs to be amplified in both E. coli and yeast, an E. coli-yeast shuttle vector that contains both an E. coli origin of replication and a yeast origin of replication (fig. 11) had to be used.
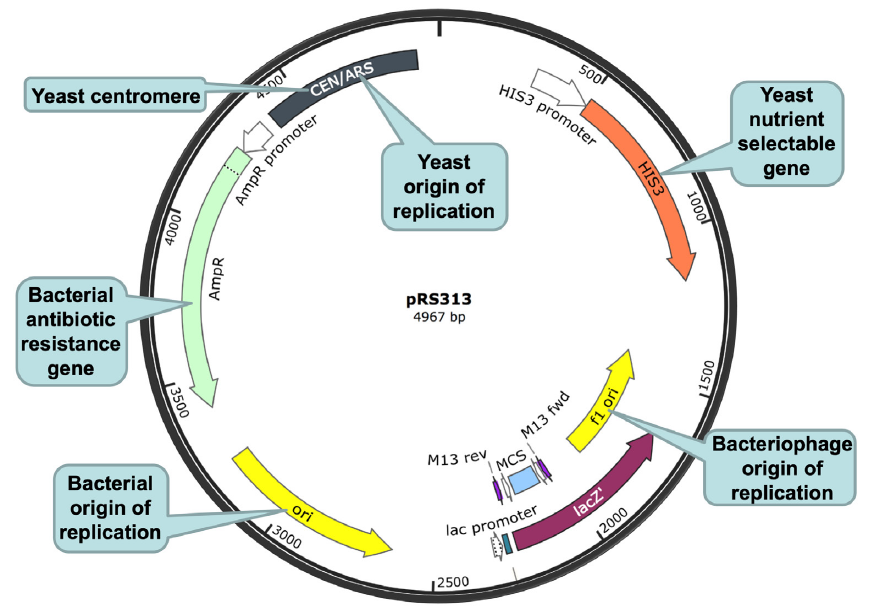
Fig. 11. An E. coli-yeast shuttle vector. The E. coli-yeast shuttle vector is shown as a black circle, with a circular ruler. The numbers on the ruler indicate the number of base pairs from the base pair at the 12 o’clock position. A few important segments of the vector are highlighted, including (clockwise, starting from the 11 o’clock position of the shuttle vector): (1) a yeast origin of replication (autonomously replicating sequence (ARS)) and a yeast centromere (cen) (gray), (2) a yeast nutrient selectable gene HIS3 (orange), (3) a bacteriophage origin of replication (f1 ori, yellow), (4) a lacZ gene (purple), (5) a multiple cloning site (MCS, light blue) located within the lacZ gene, (6) a bacterial origin of replication (ori, yellow), and (7) a bacterial antibiotic resistance gene ampR (green). The yeast origin of replication ARS is necessary for the plasmid, thus the cloned DNA, to be replicated and propagated in yeast. The yeast centromere CEN is necessary for the stability and segregation of the plasmid in yeast. The bacterial origin of replication is necessary for the plasmid, thus the DNA being cloned, to be replicated and propagated in bacteria. Both lacZ and ampR are under the control of bacterial promoters that can be recognized by the bacterial transcription machinery, thus, can be transcribed and translated in bacteria. In contrast, the yeast selectable gene HIS3 is under the control of a yeast promoter that can be recognized by the yeast transcription machinery, thus, HIS3 can be transcribed and translated in yeast. Note that ampR will not be transcribed in yeast and HIS3 will not be transcribed in bacteria because their promoters will not be recognized by the transcription machineries of the corresponding host cells. f1 ori: f1 origin of replication from an f1 phage, to enable single stranded replication and packaging into phage particles. Plasmid map is adapted from SnapGene (http://www.snapgene.com/resources/plasmid_files/yeast_plasmids/pRS313/). Used with permission.
To overcome the barriers between bacterial and eukaryotic transcription and translation machineries, the bacterial selectable genes (usually antibiotic-resistance genes) were placed under the control of a bacterial gene promoter and the yeast selectable genes (usually nutrient-selectable genes) were placed under the control of a yeast gene promoter, so that the selectable marker genes can be recognized by the transcription and translation machinery of the corresponding host organisms and be transcribed and translated. The selectable genes are necessary for the identification and isolation of the cloned DNA.
Also, because of the barriers between bacterial and eukaryotic transcription and translation machineries and difference in coding and decoding strategies, the cloned bacterial genome that was fully assembled and amplified in yeast had to be transferred into a bacterial host cell to be “activated.”
Even after all these, the initial cloned genome was not able to generate a self-replicating synthetic bacterial cell because one base pair in one of M. mycoides hundreds of essential genes was missed at the beginning of the cloning process and the mistake escaped detection along the way. The missed base pair had to be added back manually to enable the synthetic cell to survive and reproduce. For a more detailed description of what the Venter’s team did and the obstacles they had to overcome during their creation of their synthetic bacterium, the readers are referred to (Tan and Stadler 2020).
Conclusion
The inheritable genetic information that determines life or death of all living beings is not only encrypted but is encrypted in an organism-specific manner. Different organisms have to use their own unique cryptographic keys that are made of their own RNAs and proteins present in their own cells and those that they can make themselves using their own molecular machineries to decipher their genomes. This creates discontinuities in between bacteria, archaea, and eukaryotes. The discontinuities can be overcome during molecular cloning using specifically- engineered cloning vectors, but not by the organisms whose DNA is cloned, apart from human intellects. Therefore, the organism-specific genetic information coding and decoding systems challenge the popular belief that life came from non-life naturally and that all organisms are connected via a big evolutionary tree of life.
References
Chen, Xizi, Yilun Qi, Zihan Wu, Xinxin Wang, Jiabei Li, Dan Zhao, Haifeng Hou et al. 2021. “Structural Insights into Preinitiation Complex Assembly on Core Promoters.” Science 372, no. 6541 (30 April). https://www.science.org/doi/10.1126/science.aba8490.
Chen, Xizi, Xiaotong Yin, Jiabei Li, Zihan Wu, Yilun Qi, Xinxin Wang, Weida Liu, and Yanhui Xu. 2021. “Structures of the Human Mediator and Mediator-Bound Preinitiation Complex.” Science 372, no. 6546 (6 May). https://www.science.org/doi/epdf/10.1126/science.abg0635.
Gibson, Daniel G., John I. Glass, Carole Lartigue, Vladimir N. Noskov, Ray-Yuan Chuang, Mikkel A. Algire, Gwynedd A. Benders et al. 2010. “Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome.” Science 329, no. 5987 (20 May): 52–56.
Girbig, Mathias, Agata D. Misiaszek, Matthias K. Vorländer, Aleix Lafita, Helga Grötsch, Florence Baudin, Alex Bateman, and Christoph W. Müller. 2021. “Cryo-EM Structures of Human RNA Polymerase III in its Unbound and Transcribing States.” Nature Structural and Molecular Biology 28, no. 2 (February): 210–219.
Gribaldo, Simonetta, Anthony M. Poole, Vincent Daubin, Patrick Forterre, and Céline Brochier-Armanet. 2010. “The Origin of Eukaryotes and Their Relationship with the Archaea: Are We at a Phylogenomic Impasse?” Nature Reviews Microbiology 8, no. 10 (16 September): 743–852.
Koonin, Eugene V. 2015. “Origin of Eukaryotes from within Archaea, Archaeal Eukaryome and Bursts of Gene Gain: Eukaryogenesis Just Made Easier?” Philosophical Transactions of the Royal Society B-Biological Sciences 370, no. 1678 (26 September). https://royalsocietypublishing.org/doi/pdf/10.1098/rstb.2014.0333.
Labroussaa, Fabien, Anne Lebaudy, Vincent Baby, Geraldine Gourgues, Dominick Matteau, Sanjay Vashee, Pascal Sirand-Pugnet, Sébastien Rodrigue, and Carole Lartigue. 2016. “Impact of Donor-Recipient Phylogenetic Distance on Bacterial Genome Transplantation.” Nucleic Acids Research 44, no. 17 (August): 8501–8511.
Liu, Bin, Chuan Hong, Rick K. Huang, Zhiheng Yu, and Thomas A. Steitz. 2017. “Structural Basis of Bacterial Transcription Activation.” Science 358, no. 6365 (November 17): 947–951.
Matuscak, Scott T., and Change Laura Tan. 2016. “Who are the Parents of Mycoplasma Mycoides JCVI-syn1.0?” BIO-Complexity 2016, no. 2: 1–5. https://bio-complexity.org/ojs/index.php/main/article/view/BIO-C.2016.2/BIO-C.2016.2.
Nevers, Yannis, Arnaud Kress, Audrey Defosset, Raymond Ripp, Benjamin Linard, Julie D. Thompson, Olivier Poch, and Odile Lecompte. 2019. “OrthoInspector 3.0: Open Portal for Comparative Genomics.” Nucleic Acids Research 47, no. D1: D411–D418. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6323921/.
Rochette, Nicolas C., Céline Brochier-Armanet, and Manolo Gouy. 2014. “Phylogenomic Test of the Hypotheses for the Evolutionary Origin of Eukaryotes.” Molecular Biology and Evolution 31, no. 4 (April): 832–845.
Sadian, Yashar, Florence Baudin, Lucas Tafur, Brice Murciano, Rene Wetzel, Felix Weis, and Christoph W. Müller. 2019. “Molecular Insight into RNA Polymerase I Promoter Recognition and Promoter Melting.” Nature Communications 10, no. 1 (5 December): 5543.
Schmitt, Emmanuelle, Pierre-Damien Coureux, Ramy Kazan, Gabrielle Bourgeois, Christine Lazennec-Schurdevin, and Yves Mechulam. 2020. “Recent Advances in Archaeal Translation Initiation.” Frontiers in Microbiology 11: (September) 584152. https://www.frontiersin.org/articles/10.3389/fmicb.2020.584152/full.
Sehnal, David , Alexander S. Rose, Jaroslav Koča, Stephen K. Burley, and Sameer Velankar. 2018. “Mol*: Towards a Common Library and Tools for Web Molecular Graphics.” MolVA ‘18 Proceedings of the Workshop on Molecular Graphics and Visual Analysis of Molecular Data: 29–33. https://doi.org/10.2312/molva.20181103.
Tan, Change Laura. 2015. “Using Taxonomically Restricted Essential Genes to Determine Whether Two Organisms Can Belong to the Same Family Tree.” Answers Research Journal 8 (November 4): 413–435. https://assets.answersingenesis.org/doc/articles/pdf-versions/arj/v8/taxonomically_restricted_genes_family_tree.pdf.
Tan, Change Laura. 2017. “Holistic Study of Whole Genomes.” Journal of Genome 1, no. 1: 1000e102. https://www.omicsonline.org/open-access/holistic-study-of-whole-genomes.pdf.
Tan, Change. 2016. “Big Gaps and Short Bridges: A Model for Solving the Discontinuity Problem.” Answers Research Journal 9 (6 July): 149–162. www.answersingenesis.org/arj/v9/discontinuity_problem.pdf.
Tan, Change Laura. 2022a. “Facts Cannot be Ignored When Considering the Origin of Life #1: The Necessity of Bio-monomers Not to Self-Link for the Existence of Living Organisms.” Answers Research Journal 15 (9 March): 25– 29. https://answersresearchjournal.org/genetics/necessity-of-bio-monomers/.
Tan, Change Laura. 2022b. “Facts Cannot be Ignored When Considering the Origin of Life #2: Challenges in Generating the First Gene-encoding Template DNA or RNA.” Answers Research Journal 15:31–48.
Tan, Change L., and Rob Stadler. 2020. The Stairway To Life: An Origin-Of-Life Reality Check. Evorevo Books.
Tan, Change, and Jeffrey P. Tomkins. 2015a. “Information Processing Differences Between Archaea and Eukaraya— Implications for Homologs and the Myth of Eukaryogenesis.” Answers Research Journal 8 (18 March): 121–141. https://answersingenesis.org/biology/microbiology/information-processing-differences-between-archaea-and-eukarya/.
Tan, Change, and Jeffrey P. Tomkins. 2015b. “Information Processing Differences Between Bacteria and Eukarya— Implications for the Myth of Eukaryogenesis.” Answers Research Journal 8 (25 March): 143–162. https://answersingenesis.org/biology/microbiology/information-processing-differences-between-bacteria-and-eukarya/.
Venter, Craig 2010. “Watch me unveil ‘synthetic life’.” TED (Technology, Entertainment and Design). http://www.ted.com/talks/craig_venter_unveils_synthetic_life.
Vesteg, Matej, and Juraj Krajcovic. 2011. “The Falsifiability of the Models for the Origin of Eukaryotes.” Current Genetics 57, no. 6 (December): 367–390.
Williams, Tom A., Peter G. Foster, Cymon J. Cox, and T. Martin Embley. 2013. “An Archaeal Origin of Eukaryotes Supports Only Two Primary Domains of Life.” Nature 504 no. 7479 (12 December): 231–236.
Zhao, Haiyan, Natalie Young, Jens Kalchschmidt, Jenna Lieberman, Laila El Khattabi, Rafael Casellas, and Francisco J. Asturias. 2021. “Structure of Mammalian Mediator Complex Reveals Tail Module Architecture and Interaction With a Conserved Core.” Nature Communications 12, no. 1 (1 March): 1355.