The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
In a previous 2016 study aligning Sanger-style chimpanzee genomic trace reads (mean length = 704 bases) to the human genome, it was determined that chimpanzee DNA was not more than 85% similar to human. To further investigate the issue of human-chimpanzee genome similarity using higher quality DNA sequence, 18,000 de novo assembled contigs (constructed with Sanger style reads, Illumina short reads, and PacBio long reads) downloaded from NCBI having an average length of 30,913 bases were queried against the human genome using the BLASTN algorithm with gap extension. The alignments averaged 10,508 bases in length with a nucleotide identity of 84%. The contigs were also queried against the panTro4 and panTro5 versions of the chimpanzee genome yielding alignment identities of 92% and 100%, respectively. Results from this study not only negate the concept of the 98.5% DNA similarity myth, but highlight the extremely flawed and humanized nature of the panTro4 version of the chimpanzee genome and its predecessors that are widely used to support the human evolution paradigm.
Introduction
One of the chief problems with all versions of the chimpanzee genome prior to PanTro6, is that they were not constructed through the use of an accurate integrated physical-genetic map and its corresponding genomic resources in a systematic fashion like the human genome and other key model animal genomes (Tomkins 2011). Instead, short DNA sequences generated by the sequencing machinery (known as trace reads) largely produced through a whole genome shotgun approach were assembled onto the human genome using it as a reference scaffold (Mikkelsen et al. 2005; Prado-Martinez et al. 2013; Tomkins 2011). This was done not only out of convenience and a lack of available resources, but the dogmatic evolutionary presupposition that humans evolved from apes and shared a common ancestor with chimpanzees about 3 to 6 million years ago.
Another serious potential problem with earlier versions of the chimpanzee genome is the distinct possibility of human DNA contamination that would also contribute to the development of a more humanized assembly. In a previous study by this author, the first group of Sanger-style trace read data sets produced in the chimpanzee genome project during the years 2002 to 2004 that formed the basis for the initial versions of the chimpanzee genome were on average 6% more identical to human than those produced later in the project during the years 2005 to 2011 (Tomkins 2016).
The problem of human DNA contamination in public databases is a valid concern. In 2011, a scientifically disturbing study was published in which researchers evaluated 2749 non-primate public DNA databases and determined that 492 were contaminated with human sequence at levels of up to 10% (Longo, O’Neill, and O’Neill 2011). The contaminated DNA databases represented species including bacteria, plants, and fish. Ape and monkey databases were not screened, leaving the question pending as to how much human DNA contamination may be present in non-human primate genomes. More recently, another research study was done investigating this issue in which the presence of contaminating human DNA in non-primate public databases was found to persist (Kryukov and Imanishi 2016). The authors of the report stated, “We recommend that existing contaminated genomes should be revised to remove contaminated sequence, and that new assemblies should be thoroughly checked for presence of human DNA before submitting them to public databases.”
It is also well known that archaic human DNA sequencing projects such as Neandertal have been pestered with the problem of modern human DNA contamination that have led to the development of much stricter laboratory precautions (Skoglund et al. 2014; Thomas and Tomkins 2014). Nevertheless, modern human DNA contamination is a standard problem in the first generation of published ancient DNA studies (Noonan 2010; Skoglund et al. 2014; Skoglund, Thomas, and Tomkins 2014).
While the problem of human DNA contamination in the chimpanzee genome has never been addressed by the secular community, researchers have recently openly acknowledged sequence assembly problems stating, “the higher-quality human genome assemblies have often been used to guide the final stages of nonhuman genome projects, including the order and orientation of sequence contigs and, perhaps more importantly, the annotation of genes” and “This bias has effectively “humanized” other ape genome assemblies” (Kronenberg et al. 2018). Even with a more recent version of the chimpanzee genome (PanTro5) that used a hybrid approach of next generation sequencing technologies, including PacBio long reads, the resulting contiguous pieces of de novo assembled DNA sequence were still oriented and aligned onto the human genome as a reference (Kronenberg et al. 2018; Kuderna et al. 2017).
At the time of this publication, a new version of the chimpanzee genome has been announced (PanTro6) that was assembled completely de novo without the use of a human as a reference scaffold (Kronenberg et al. 2018). According to correspondence with UCSC genome browser staff at the time of this report, “The panTro6 assembly has not yet been reviewed by our Quality Assurance team” and is not available for public download. However, LASTZ alignments with the human genome have been performed and are available for download (ftp://hgdownload.cse.ucsc.edu/goldenPath/hg38/vsPanTro6/). LASTZ is a large-scale genome alignment tool that can efficiently align chromosomal or genomic sequences millions of nucleotides in length.
Queen Mary University of London evolutionary geneticist, Richard Buggs, recently performed an analysis of the UCSC LASTZ results and reported, “The percentage of nucleotides in the human genome that had one-to-one exact matches in the chimpanzee genome was 84.38%” (Buggs 2018). Not only do these LASTZ PanTro6 results fit well with a previous report by Tomkins (2016) in which it was determined that the chimpanzee genome could be no more than 85% similar to human, but these results also match closely with data described below in this present study.
Interestingly, Buggs also calculated the amount of sequence that was unalignable between human and chimpanzee stating, “4.06% had no alignment to the chimp assembly.” Assuming that the genome sizes between human and chimpanzee are similar, when the non-alignable sequence data is combined with the alignment data (Buggs 2018), the current level of overall human-chimpanzee genome similarity can now be estimated at about 80%.
Despite the recent improvements with the PanTro5 and PanTro6 versions of the chimpanzee genomes, no objective reassessment of human chimpanzee genome similarity has been forthcoming from the secular research community outside of the recent internet post by Buggs (2018), which at the time of this report, has received no credible challenge or rebuttal.
In an attempt to get around the bias presented by the humanized chimpanzee genome assembly issue, in a previous study, I sampled 25,000 unassembled trace reads at random from each of the 101 Sanger-style trace read data sets that provided the foundation for the initial versions of the chimpanzee genome (Tomkins 2016). As a follow-up to this previous research, and in an attempt to use higher quality, less contaminated (with human DNA), and longer sequences, 18,000 publicly available de novo assembled contigs combining Sanger-style reads, Illumina short reads, and PacBio long reads were queried against the human genome using the BLASTN algorithm with gap extension.
Materials and Methods
Assembled chimpanzee sequencing contigs with accession numbers AACZ0400000-AACZ04072784 were downloaded from the European Nucleotide Archive (www.ebi.ac.uk). According to the assembly release notes, these de novo assembled contigs represented a pure ‘Clint’ version of the chimpanzee genome generated from a 6-fold coverage of Sangerstyle reads, 55-fold coverage of Illumina overlapping paired 250bp length reads and a 9-fold coverage of PacBio long single molecule reads.
18,000 contigs were chosen at random and queried against the GRCh37.71 version of the human genome and the PanTro4 and PanTro5 versions of the chimpanzee genome using BLASTN v2.2.31 with the following parameters: evalue 0.1, word_size 11, outfmt 10, qseqid, qstart, qend, mismatch, gapopen, pident, nident, length, qlen, max_target_seqs 1, max_hsps 1, dust no, soft_masking false, perc_identity 50, gapopen 3, gapextend 3, num_threads 10. Individual BLAST Jobs were run in parallel on two Intel Xeon E5 v2/Core i7servers each having 40 logical cores and 380 GB RAM. Resulting BLASTN output CSV format files and FASTA format sequence files were analyzed for a variety of basic statistical parameters using a Python script written by this author. CSV output files and Python scripts used in this study are available at GitHub (https://github.com/jt-icr/chimp_contigs).
Results
Contig size statistics
An assessment of the 18,000 chimpanzee sequencing contigs used in this study revealed the mean length was 30,913 bases, median length was 1832 bases, minimum length was 208 bases and the maximum was the 2,729,125 bases. A graphical depiction of contig distribution by size is shown in fig. 1. A majority of the contigs (90%) were less than 50,000 bases in length and 96% were less than 250,000 bases.
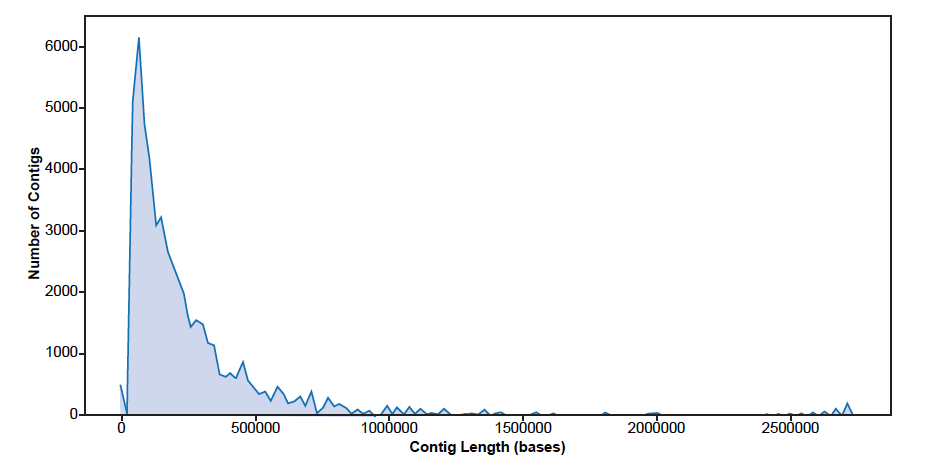
Fig 1. Distribution of de novo assembled chimpanzee sequencing contigs by size.
Comparison to Human
The main finding of significance to the issue of alleged common ancestry between humans and chimpanzees is the fact that the average alignment identity was only 84% (table 1). Despite the gap extension parameters being quite liberal, the average mean alignment length was only 10,509 bases as a result of the algorithm hitting a gap that was too large for it to traverse. Thus, only about one-third of each chimpanzee contig on average could be aligned to the human genome as the best hit. These data obviously exclude the less alignable portions of the contigs as well as those regions that would be completely unalignable. Thus, the overall identity of the chimpanzee genome compared to human would actually be significantly lower than 84%.
| Target database | |||
|---|---|---|---|
| Human GRCh37.71 | Chimp PanTro4 | Chimp PanTro5 | |
| Mean alignment identity (percent) | 84.39 | 91.91 | 100.0 |
| Median alignment identity (percent) | 88.40 | 91.97 | 100.0 |
| Minimum alignment identity (percent) | 64.13 | 66.32 | 95.76 |
| Maximum alignment identity (percent) | 100.0 | 100.0 | 100.0 |
| Average alignment length (bases) | 10,509 | 10,699 | 30,544 |
| Median alignment length (bases) | 1,370 | 1,705 | 1,832 |
| Minimum alignment length (bases) | 30 | 26 | 208 |
| Maximum alignment length (bases) | 342,162 | 451,828 | 2,729,125 |
| Mean percent of total sequence aligned | 33.97 | 34.60 | 98.80 |
| Number of hits | 17,989 | 17,995 | 17,999 |
| Average hit frequency | 99.94 | 99.97 | 99.99 |
Comparison to PanTro4
The PanTro4 assembly of the chimpanzee genome has been the version most commonly used in recent years to support an alleged common ancestry with human. However, both this author (Tomkins 2011, 2016) and more recently, authors of the new de novo assembled PanTro6 version of the chimpanzee genome have asserted that past versions of the chimpanzee genome have been “humanized” (Kronenberg et al. 2018). This is especially true for the PanTro4 version and its predecessors.
The main finding of significance to the issue of humanization of the chimpanzee genome is the fact that the average mean alignment identity of the de novo assembled chimpanzee contigs was only 91% when queried against the PanTro4 assembly, not 100% as would be expected if the chimpanzee genome was an accurate representation (table 1). In fact, some regions had alignment identities lower than 70% with a minimum as low as 66%. The alignments were so poor that the average mean alignment length of only 10,699 bases was not much better than that achieved using human as a target database. Thus, only about one-third of each chimpanzee contig on average could be aligned to the PanTro4 version of the chimpanzee genome as the best hit. These poor alignments indicate that the humanization of the chimpanzee genome was extremely severe and heavily biased towards an evolutionary outcome.
Comparison to PanTro5
The improved PanTro5 version of the chimpanzee genome was released and published recently using the same de novo assembled sequencing contigs accessed in this study (Kuderna et al. 2017). However, the contigs were still oriented and merged onto the human genome as a reference assembly. Much of the humanization had been alleviated at more of a micro level, although the overall assembly was likely still significantly humanized as noted in the publication of the recent PanTro6 version (Kronenberg et al. 2018). Nevertheless, the PanTro5 version of the chimpanzee genome served as a good target database for an experimental control treatment in this study.
Alignments for the chimpanzee sequencing contigs onto the PanTro5 assembly had a nucleotide identity of 100%, indicating that the BLASTN algorithm was functioning properly and that the PanTro5 version of the chimpanzee genome was relatively accurate, at least on a local alignment level. In good agreement with the identical alignments were the average alignment lengths of 30,544 bases being nearly similar to the average contig length of 30,913 bases. The average 369 base discrepancy in unalignable DNA may very well be due to the merging of the contigs onto the human reference in PanTro5, slightly truncating the average alignment lengths.
Results from querying the chimpanzee sequencing contigs on the PanTro5 version of the chimpanzee genome served as a strong control measure and validated the overall experiment. The results also highlight the distinct difference in assembly quality between the PanTro4 and PanTro5 versions of the chimpanzee genome, illustrating the highly humanized evolutionary bias of PanTro4.
Summary and Conclusion
Early versions of the chimpanzee genome assembly suffer from two major problems that make it more human-like than it should be. First, chimpanzee DNA sequences from both Sanger-style sequencing and next generation short-read sequencing technologies, were assembled using the human genome as a reference framework (Mikkelsen et al. 2005; Prado-Martinez et al. 2013). Second, given the fact that significant levels of human DNA exist in non-primate databases due to laboratory and worker contamination (Longo, O’Neill, and O’Neill 2011), the potential for human DNA in the pre-assembled chimpanzee sequencing reads is highly probable. This contention of possible human DNA contamination was supported by results in a study done by this author evaluating a sampling of 25,000 reads from each of the 101 Sanger-style trace read data sets used to produce initial versions of the chimpanzee genome (Tomkins 2016). However, the recent merging of Sanger-style reads and Illumina short-reads with PacBio long-reads along with improved lab techniques has likely removed human DNA contamination as a significant issue in the PanTro5 and the new PanTro6 versions of the chimpanzee genome.
When blasting chimpanzee trace reads onto an allegedly accurate representation of the chimpanzee genome, one would expect alignment identities of 100% as was achieved in this study using the PanTro5 assembly as a control. However, the average alignment identity (excluding all non-hitting sequence) for the chimpanzee contigs in this study onto the PanTro4 version of the chimpanzee genome, was only 91.9% combined with an alignment length of only about one-third of the query contig sequence. These results strongly suggest that the early versions of the chimpanzee genome are miss-assembled and considerably more human-like than they should be. Evolutionary arguments of nearly identical DNA similarity are based on these flawed and humanized versions of the chimpanzee genome.
Perhaps the most noteworthy outcome of this current study is the fact that the alignable regions of the chimpanzee sequencing contigs were only 84.4% identical to their respective matches in the human genome. In addition, on average, only about one-third of each contig could be aligned using liberal gap extension parameters. Thus, the 84.4% nucleotide identity of the alignments is not an indicator of overall genome similarity because it does not include the regions of the contigs that are so different that they are non-alignable.
In a previous report in which Sanger-style chimpanzee trace read data sets ascertained to have reduced levels of human DNA contamination were aligned onto the human genome, it was determined that chimpanzee DNA could not be more than 85% similar to human overall (Tomkins 2016). The more comprehensive results published in this report both support and refine these earlier findings.
Most importantly, it is both fortuitous and highly noteworthy that the alignment identities achieved in this current study are extremely similar to an analysis of UCSC LASTZ human-chimpanzee (PanTro6) alignments analyzed recently by a Queen Mary University of London geneticist who reported, “The percentage of nucleotides in the human genome that had one-to-one exact matches in the chimpanzee genome was 84.38%” (Buggs 2018). The identities of the alignable DNA in this study and that obtained from LASTZ alignments are both 84% and in perfect agreement.
Furthermore, neither this study nor the LASTZ matched nucleotides account for the unalignable regions of the genome between human and chimpanzee. From the LASTZ alignment data, Buggs calculated, “4.06% had no alignment to the chimp assembly.” If this non-alignable sequence data is combined with the known alignment data (Buggs 2018), a reasonably accurate estimate of overall human-chimpanzee genome similarity would be close to 80% assuming that the genome sizes are similar.
A glaring 20% overall DNA similarity difference between the human and chimpanzee genome is an evolutionary discrepancy that cannot be dismissed. This extreme level of genetic discontinuity raises serious issues for the evolutionary myth that humans and chimpanzees share a common ancestor not more than about 3 to 6 million years ago which largely depends on a 98 to 99% DNA similarity to seem theoretically possible. The uniqueness of mankind as stated in Genesis, “So God created man in His own image; in the image of God He created him; male and female He created them,” (Genesis 1:27 (NKJV)) is now soundly confirmed by the scientific data.
References
Buggs, Richard. 2018. “How Similar Are Human and Chimpanzee Genomes?” http://richardbuggs.com/index.php/2018/07/14/how-similar-are-human-and-chimpanzee-genomes/.
Kronenberg, Zev N., Ian T. Fiddes, David Gordon, Shwetha Murali, Stuart Cantsilieris, Olivia S. Meyerson, Jason G. Underwood, et al. 2018. “High-Resolution Comparative Analysis of Great Ape Genomes.” Science 360, no. 6393: eaar6343.
Kryukov, Kirill, and Tadashi Imanishi. 2016. “Human Contamination in Public Genome Assemblies.” PLoS One 11, no. 9: e0162424.
Kuderna, Lucas F. K., Chad Tomlinson, LaDeana W. Hillier, Annabel Tran, Ian T. Fiddes, Joel Armstrong, Hafid Laayouni, et al. 2017. “A 3-Way Hybrid Approach to Generate a New High-Quality Chimpanzee Reference Genome (Pan_tro_3.0).” Gigascience 6, no. 11: 1–6.
Longo, Mark S., Michael J. O’Neill, and Rachel J. O’Neill. 2011. “Abundant Human DNA Contamination Identified in Non-Primate Genome Databases.” PLoS One 6, no. 2: e16410.
Mikkelsen, T. S., L. W. Hillier, E. E. Eichler, M. C. Zody, D. B. Jaffe, S. P. Yang, W. Enard, et al. 2005. “Initial Sequence of the Chimpanzee Genome and Comparison With the Human Genome.” Nature 437, no. 7055: 69–87.
Noonan, J. P. 2010. “Neanderthal Genomics and the Evolution of Modern Humans.” Genome Research 20, no. 5: 547–553.
Prado-Martinez, Javier, Peter H. Sudmant, Jeffrey M. Kidd, Heng Li, Joanna L. Kelley, Belen Lorente-Galdos, Krishna R. Veeramah, et al. 2013. “Great Ape Genetic Diversity and Population History.” Nature 499, no. 7459: 471–475.
Skoglund, Pontus, Bernd H. Northoff, Michael V. Shunkov, Anatoli P. Derevianko, Svante Pääbo, Johannes Krause, and Mattias Jakobsson. 2014. “Separating Endogenous Ancient DNA from Modern Day Contamination in a Siberian Neandertal.” Proceedings of the National Academy of Sciences USA 111, no. 6: 2229–2234.
Thomas, B., and J. Tomkins. 2014. “How reliable are genomes from ancient DNA?” Journal of Creation 28, no. 3: 92–98.
Tomkins, Jeffrey P. 2011. “How Genomes are Sequenced and Why it Matters: Implications for Studies in Comparative Genomics of Humans and Chimpanzees.” Answers Research Journal 4: 81–88. https://answersingenesis.org/genetics/dna-similarities/how-genomes-are-sequenced-and-why-it-matters/.
Tomkins, Jeffrey P. 2016. “Analysis of 101 Chimpanzee Trace Read Data Sets: Assessment of Their Overall Similarity to Human and Possible Contamination with Human DNA.” Answers Research Journal 9: 294–298. https://answersingenesis.org/genetics/dna-similarities/analysis-101-chimpanzee-trace-read-data-sets-assessment-their-overall-similarity-human-and-possible/.