Research conducted by Answers in Genesis staff scientists or sponsored by Answers in Genesis is funded solely by supporters’ donations.
Abstract
Microbes form a life-sustaining organosubstrate on earth and contribute to our understanding of geology, ecology, and biology. Creation microbiologists are actively involved in research and publication, but a field of creation microbiology has not yet been formally established. This paper represents a first attempt to organize the field of creation microbiology by providing an overview of the general classification of microbes and their functions in the biospher.
Keywords: organosubstrate, creation, bacteria, virus, fungi, protist, algae, microbiology
Introduction
It is the goal of this paper to provide an introduction to the field of creation microbiology. Microbiology is the study of microbes which are typically microscopic organisms which cannot be readily seen by the naked eye. Microbes are the most abundant creature on earth. It is estimated that there are 109 times more microbes on earth than there are stars in the universe.1 Because microbes cause disease we tend to think of them as a source of natural evil, yet many of these creatures have now been recognized as performing life-sustaining functions as part of an organosubstrate on earth (Francis 2003). Thus, the discipline of microbiology has the potential to contribute greatly to our understanding of living organisms and the development of a biology creation model. However, few papers have been published by creationists in this field. Therefore, as a first approach towards the establishment of a field of creation microbiology we provide an introductory overview of the general taxonomical categories of microbes and discuss their diversity, abundance, and function in the biosphere.
Abundance and Diversity in Form, Species, and Habitat of each Microbe Type
Microbes can be classified broadly according to their cell structure as eukaryotic or prokaryotic, (Anderson 1980; Bergman 1998) or lack of cell structure as in the case of viruses or prions. Prokaryotes are more generally referred to as bacteria. The taxonomy of eukaryotic microbes is complex and reflects their wide disparity in form and function. We will discuss the diversity of cellular microbes within the context of their classification as prokaryotic or eukaryotic keeping in mind that in nature microbes are not segregated by class but instead intimately associate together forming a living organosubstrate matrix (Francis 2003). We will begin by discussing viruses which exist on the edge of life and cellularity.
Non-cellular microbes: viruses
Viruses2 are abundant in all ecosystems and within all living organisms. Conservative estimates put the total number of viral species on earth at 400,000. It is very possible that the number of virus species could be greater than 10 million because every bacterial species is predicted to possess at least one unique phage symbiont (table 1) (Chapman 2005).3 Roughly, 2,000 species of viruses have been documented, however over 3,000 unassigned viral nucleic acid sequences have been submitted to sequence databases (Fauquet and Fargette 2005).
Viruses are typically smaller than cells but exhibit a remarkable diversity of genomic structure, size and morphology for creatures which possess fewer complex structures than cellular organisms. Viruses have traditionally been assumed to be several orders of magnitude smaller than the smallest cells. Recently, the discovery of the giant Mimivirus, has challenged this assumption. This giant virus resides in the protozoan Acanthamoeba polyphaga and is physically larger than small bacteria; its genome at 1.2 Mb is 2.5 times larger than the smallest bacterial genomes. Remarkably, the Mimivirus can also be infected with small viruses prompting the proposal of a new category of viruses; the virophages (La Scola et al 2008). Virophages are predicted to shuttle nucleic acids among large viruses (La Scola et al 2008).
| Organism | Number of known species | Predicted number of species | Size range (µm) | Genome size range, C-value (pg) |
|---|---|---|---|---|
| eubacteria | 9000 | 10,000,000 | 0.1–750 | 10-3–10-2 |
| archea | 300 | ? | 1–? | 10-3–10-2 |
| fungi | >74,000 | 1,500,000 | 5–5.6 × 109 | 10-2–1.0 |
| algae | >34,000 | 400,000 | 1–6 × 107 | 10-2–10-1 |
| protists | >42,000 | 400,000 | 2–200 | 10-2–103 |
| viruses | >2000 | 10,000,000 | 0.005–0.4 | 10-6–10-2 |
The elucidation of the role of viruses in nature has focused primarily on their disease causing abilities (Francis 2003). A creationist perspective of viruses demonstrates that the beneficial functions of viruses can be organized into at least four categories, 1) population control; the population dynamics of many bacteria is influenced by phages, for instance, prevention of over-population of cholera bacteria in waterways is regulated by bacteriophages (Jensen et al. 2006), 2) mobile delivery agents and genetic elements; viruses deliver proteins, lipids, and nucleic acids to many kinds of cells and organisms with the potential to alter cells and tissues epigenetically and genetically (Gruenke, Francis and Wood 2004; Lindell et al. 2004), for instance, viruses are such potent genetic modulators of marine algae that they are predicted to influence global biogeochemical cycles and weather patterns (Wilson, Van Etten and Allen 2008). Viruses may also be involved in speciation events in metazoans (Wood 2002, 2003), 3) participation in immune defense; viruses have played a role in protecting organisms like the American Chestnut tree from pathogens (Jaynes and Elliston 1980) or muting the immune system, for example, in the vicinity of the human embyro (Villareal 1997), 4) biochemical switches; viral factors interact with many different biochemical pathways, and in many cases they can turn a pathway on or off (Gruenke, Francis and Wood 2004). In addition viruses have been used in medicine to promote cancer treatments via oncolysis (Kim 2008). Undoubtedly many other beneficial viral functions in nature will be uncovered as more viral species are discovered and characterized.
Prokaryotic microbes: bacteria
Bacteria as a group are species rich and display a wide range of habitat; they are found in the atmosphere, deep within the earth and oceans, and on and within every living creature. Bacteria can be broadly classified into two groups, the eubacteria and the archaea. Archaea are bacteria which live in extreme environments. It is estimated that 9,000 species of eubacteria have been identified. In contrast, less than 300 species of the more recently discovered archaea have been identified. Microbiologists have surveyed the bacterial DNA content of soils, waterways and the human body; from this analysis they estimate that greater than 10 million species of bacteria may exist in the biosphere. Some ecosystems are extremely densely populated. For instance, the human gastrointestinal tract, which may be the most densely concentrated ecosystem on earth, contains over 5,000 species of bacteria and these bacteria are part of a microbial community of 10–100 trillion organisms (Dethlefsen et al. 2008). In addition, bacteria are also found to be in the highest concentration at interfaces between major parts of the biosphere, that is, the interface between the lithosphere and hydrosphere, or the hydrosphere and atmosphere (Curtis, Sloan, and Scannell 2002). This is consistent with the idea that they are major purveyors of biogeochemical cycles.
Interestingly, laboratory specimens of bacteria show only few cellular morphological forms, however this belies the fact that there is greater genetic diversity among bacteria than there is among mammals or possibly even vertebrates. For instance, remarkably, two different strains of E. coli, can differ by more than a third of their genomes (over 2000 genes; Lane 2007). Lateral gene transfer has been proposed as a hypothesis for this wide disparity in genomes, however despite the possible prevalence of lateral gene transfer which is a process which adds DNA to the genome, bacterial genomes have remained small compared to eukaryotic genomes. (Lane 2007; Ochman, Lawrence and Groisman 2000). For instance, there are no known bacterial genomes greater than 10 Mb (megabases); eukaryotic genomes are larger by up to 4 orders of magnitude. In fact, the largest known genome is 670,000 Mb possessed by the eukaryotic microbe, Amoeba dubia. Strikingly this genome is 200 times larger than the human genome (Lane 2007).
In addition, even though bacteria show little diversity in cellular forms, most of this work has been based on observations of laboratory strains of bacteria which are raised in sterile environments. Many bacteria and microbes display a greater diversity in morphology when they are observed in their natural setting (fig. 1). In fact, many are pleomorphic, appear plant-like and are involved in complex community interactions.
Although we often think of bacteria as causing disease, overall, bacteria provide a life-supporting function on earth by their participation in all biogeochemical cycles (Francis 2003, 2008). Bacteria also possess design features which allow them to interface with and transfer nutrients to living organisms.
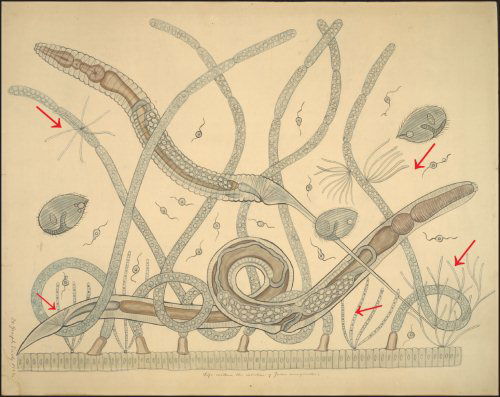
Fig. 1. The intestinal microbial community of the millipede Julus marginatus drawn as a teaching chart by Joseph Leidy. Margulis et al. (1998) have shown that the fine filamentous growths are the bacterium Bacillus cereus, which displays filamentous growth in situ (arrows). Joseph Leidy referred to these thin filaments as Arthromitus in 1849 (Leidy 1849). Note that the filamentous B. cereus attaches to the intestinal wall and to fungal hyphae. Teaching chart no. 60. Displayed as a figure in Margulis et al, 1998. Used with permission of the Academy of Natural Sciences of Philadelphia, Ewell Sale Stewart Library and the Albert M. Greenfield Digital Imaging Center for Collections.
Eukaryotic microbes
Eukaryotic microbes are diverse in size, morphology, ecological function and lifestyle. Eukaryotic microbes play major roles as part of the life-supporting biomatrix. For instance, a majority of the carbon obtained by organisms is obtained (fixed) from the atmosphere by these microbes. Eukaryotic microbes have been traditionally categorized according to their cell surface morphologies and niche occupation, however, because of the recent focus on analysis of rDNA sequences their classification has undergone dramatic shifts in the last few years. Currently, all eukaryotes are classified in six supergroups (see table 2). This classification system is somewhat cumbersome and incomplete because it is currently undergoing revision and most likely will require constant updating. For instance, a recent survey of the sea surface waters in several of the world’s oceans showed a surprising abundance of picoeukaryotes (small eukaryotic microbes !!LESS THAN SIGN!! 3mm in size) (Massana and Pedros-Alio 2008). Eukaryotic microbes in these habitats have tended to be overlooked because microbiologists have focused on prokaryotes. To the surprise of the investigators an abundance of picoeukaryotes were discovered in these waters with representatives in all the supergroups except for the ameobozoa. Fascinatingly, a significant number of picoeukaryotes were discovered which did not fit in any of the six supergroups (Massana and Pedros-Alio 2008).
| Supergroups | Representatives |
|---|---|
| Archaeplastida (Plantae) |
Red algae Green algae Plants |
| Chromalveolata | Ciliates Apicomplexa |
| Excavata | Euglenozoa Diplomonads Parabasalids |
| Opishtonokonta | Animals Fungi Choanoflagellates |
| Rhizaria | Foraminifera Cercomonads Euglyphids |
| Amoebozoa | Amoeba Slime molds |
For the sake of simplicity we will consider the species diversity of eukaryotic microbes within the traditional common categories of Fungi and Protists. In addition, we have further sub-divided the Protist group into the photosynthetic protists and the nonphotosynthetic protists.4
Fungus
Fungi differ from bacteria in that they are eukaryotic in cell organization and they differ from algae because they do not perform photosynthesis. It is estimated that 74,000–120,000 fungi have been documented representing over 5% of all described species on earth. It is estimated that there are over 1.5 million species thriving in the biosphere (Burnett 2003; Hawksworth and Mueller 2005). Studies which examined the frequency of fungal association with plants suggest that there are a minimum of 600,000 fungal species.
| Living Vascular Plants |
|---|
| Biotrophs and necrotrophs of leaves, stems, flowers, fruits, seeds, roots, etc. |
| Commensals on bark and leaves (especially lichen-forming fungi) |
| Endophytes of leaves, stems, bark, and roots |
| Secondary colonizers of dead attached tissues and leaf spots, etc. |
| Mycorrhizas (endo-, ecto-, ericoid, orchid, etc.) |
| Leaf surfaces |
| Nectar |
| Resin |
| Dead Vascular Plants |
| Saprobes on wood, bark, and litter |
| Burnt plant tissues |
| Saprobes on submerged and inundated plants |
| Pollen in water samples |
| Nonvascular Plants |
| Algae (marine, terrestrial, and freshwater) |
| Bryophytes |
| Fungi |
| Biotrophys, necrotrophs, and saprobes of other fungi |
| Lichenicolous fungi |
| Myxomyceticolous fungi |
| Vertebrates |
| Skin, feathers, hair, bone, etc. |
| Dung |
| Nests, lairs, etc. |
| Ruminant guts |
| Fish scales and guts |
| Invertebrates |
| Biotrophs and nectotrophs |
| Arthropods exoskeletons |
| Arthropod and annelid guts |
| Nematodes |
| Insect nests |
| Rock |
| Lichens |
| Epilithic fungi |
| Endolithic fungi |
| Soil |
| Surface |
| Soil cores |
| Water |
| Foam |
| Streams, permanent and temporary ponds |
| Litter and wood immersed in sea- and freshwater |
| Plants (for example, bromeliads) |
Fungi inhabit many ecosystems and tend to be found wherever bacteria can be found (table 3). The growth of fungi tend to be noticed more often than bacteria because fungi tend to form macroscopic structures more readily than bacteria in the human environment. One macroscopic fungi which are not easily observable because they are below ground are the myccorrhizal fungi which inhabit the roots of plants. Mycorrhizae associate with 80% of all plants and are predicted to play a critical role in nutrient uptake (Van der Heijden et al. 1998; Whitfield 2007). For instance, the underground network of Mycorrhizae connects trees together and allows them to exchange nutrients; this has been referred to as a living underground Internet (Simard et al. 1997). In fact a single gram of soil can contain 100 meters of mycorrhizal filaments (Whitfield 2007). Because mycorrhizae are so efficient at helping plants obtain nutrition, over 400 plant species do not perform photosynthesis and have no chlorophyll. For example, consider the brilliantly red snow plant Sarcodes sanguinea which grows in mountainous regions on the west coast of the U.S. (fig. 2). This nonphotosynthetic plant obtains nutrition from a group of mycorrhizae fungi which may be unique to the snow plant (Kruetzer et al. 2000). Also, evidence suggests that pine trees contribute nutrients to the snow plants via the mycorrhizal connections (Bidartondo et al. 2000).

Fig. 2. The red snow plant Sarcodes Sanguinea is a member of the blueberry and cranberry family of plants (Heaths). This plant which grows in mountainous regions in the western U.S. is non-photosynthetic and obtains all its nutrition from mycorrhizae which are often associated with nearby pine tree roots. The snow plant is considered to be a representative of over 400 known “mycoheterotrophs” which obtain fixed carbon from Fungi.
Mycorrhizal diversity can be astoundingly high; 2,000 species of mycorrhizal fungi have been detected growing on Douglas Fir trees (Baxter and Dighton 2005).
Most all fungus grown on media in the laboratory display a very similar hyphae structure, which are the microscopic strands which appear like fuzzy growths seen on spoiled fruit or old bread for example. However, in situ, fungi can display unique morphologies. For example, fungi are consumed as a main source of nutrition among many insects and other invertebrates: in many cases the insects grow and “farm” the fungi (Silliman and Newell 2003). In these farms the fungal morphology is dramatically different from the typical hyphal structure observed in the laboratory (fig. 3). Fungi also demonstrate a large range of diversity of size. In fact the largest organism on earth is a fungus, Armillaria ostoyae which is found in northern forests from Oregon to Michigan (Smith, Bruhn, and Anderson 1992) with the largest recorded specimen covering 2,200 acres in Oregon.
Fungi have a great potential for genetic diversity because they demonstrate the unique property of being able to fuse their hyphae together creating heterokaryotic (a multicellular fungus which contain different nuclei) and heteroplasmic (a multicellular fungus which contains different kinds of cytoplasm and organelles) individuals. Remarkably, this genetic diversity can be observed in a single individual fungus. For example, in the Glomales (the arbuscular mycorrhizal fungi) a remarkably wide disparity of rDNA (ribosomal DNA) sequences can be detected within multiple nuclei of each individual fungal spore of a single individual fungus. What is striking is that the diversity is expressed in the rDNA sequences which are known as the most highly conserved DNA regions in all biological creatures; in fact these ribosomal sequences are widely used for taxonomy and phylogenetics (Sanders 1999). Using this criterion, the internal tissue cells of an individual Glomales could differ more than Glomales differs from another creature. This has caused some consternation among fungal geneticists and some have commented in the scientific literature:
What is clear is that evolutionary biologists and fungal geneticists are faced with a happy disconcerting puzzle. (Sanders 1999).
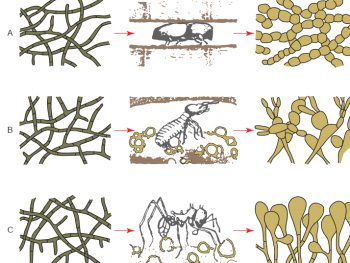
Fig. 3. Dramatic change of fungal morphology of fungi in the presence of insect farmers. The left hand side of the diagram shows fungal hyphae structures of three different fungi. In the laboratory the fungal morphologies are very similar and look identical. However, when these fungi are viewed in the insect colony nest environment a very different morphology is observed (after Batra and Batra 1967).
The major function of fungi in the environment appears to be nutrition provision. Fungi provide nutrition in ecosystems in three major ways: the decomposer/detritivore cycle, mycorrhizae and the lichen partnership. In the decomposer/detritivore cycles fungi specialize in recycling nutrients from decaying matter and are major saprophytic decomposers in the terrestrial landscape. In the forest ecosystem 90% of plant material is processed in these cycles (Ricklefs 2008). Fungi and bacteria help recover many of the nutrients in these decay cycles and fungi specialize in breaking down tough polysaccharides which few other microbes can digest (Ricklefs 2008). The hyphae of fungi are designed to penetrate dense plant material like woody stems and roots which few microbes can access (Ricklefs 2008). In this way fungi provide partially broken down organic matter which helps fuel the bacteria-based biogeochemical cycles. Fungi also participate in nutrition via symbiotic partnerships with plants (mycorrhizae) and algae (lichens).
Photosynthetic protists; the algae
Perhaps the greatest diversity in form and function at the cellular level in eukaryotes is found in the algae (fig. 4a and 4b). It is estimated that there are over 34,500 species described with 200–400,000 estimated to exist in the biosphere (Groombridge and Jenkins 2002). Algae are mainly aquatic organisms; many gametes and zoospores demonstrate swimming abilities. Reproduction methods in algae are also tremendously diverse and perhaps more diverse than any other microbe or cell type (fig. 5). Many algae can exist in moist environments and in harsh climates like the desert where they can be found in the surface cracks in stones and building structures like concrete structures and roofs. Algae can be found on the surface of plants as well as on the abundant surfaces provided by ice and snow. Algae are also one of the most intimate of all symbiotic partners, living within cells and internal spaces of many aquatic organisms where they provide nutrition by essentially harvesting solar power. For instance, the solar powered sea slug uses surgical precision to disrupt algae cells obtaining intact chloroplasts which it then inserts into sun-exposed cells on its dorsal surface (Hennigan 2008). Algae are also one of the key symbiotic partners in lichens.
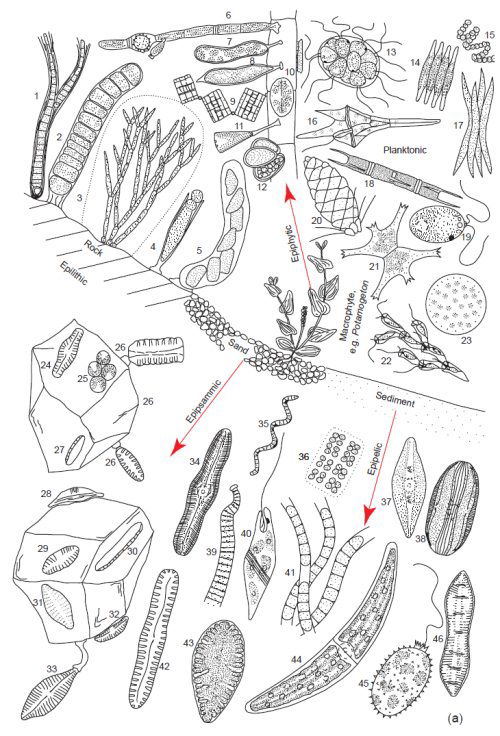
Fig. 4a. Examples of the tremendous diversity in cell form of the microscopic planktonic algae in freshwater. Epilithic = growing on rocks, Epipsammic = growing in sand, Epipelic = growing in sediments, Epiphytic = growing on plants. (Round 1981). Used with permission of Cambridge University Press.
These area all microscopic genera, a few of which may form filaments or crusts visible to the naked eye. Epilithic: 1. Calothrix, 2. Ulothrix, 3. Chaetphora, 4. Chamaesiphon, 5. Cymbella. Epiphytic: 6. Oedogonium, 7. Ophiocytium, 8. Characium, 9. Tabellaria, 10. Cocconeis, 11. Gomphonema, 12. Dermocarpa. Planktonic: 13. Pandoria, 14. Fragilaria, 15. Anabaena, 16. Ceratium, 17. Ankistrodesmus, 18. Melosira, 19. Chlamydomonas, 20. Mallonomas, 21. Staurastrum, 22. Dinobyron, 23. Cyclotella, 24 Navicula, 25 Green Unicells, 26. Opephora, 27. Nitzschia, 28. Amphora, 29. Achnanthes, 30. Nitzschia, 31. Achnanthes, 32. Cymbella, 33. Gomphonema. Epipelic: 34. Caloneis, 35. Spirulina, 36. Mesimopedia, 37. Navicula, 38. Amphora, 39. Oscillatoria, 40. Euglena, 41. Phormidium, 42. Pinnularia, 43. Surirella, 44. Closterium, 45. Trachelomonas, 46. Cymatopluera.
Algae are not known to populate many terrestrial land animals but they have been found growing on human teeth and they live symbiotically on the specially designed fur of sloths where they may provide camouflage for the sloth (fig. 6) in addition to a unique miniature ecosystem which includes microbes and insects (Aiello 1985).
Algae are a vital link in the aquatic food chain and they play a major role in the global carbon cycle; it is estimated that they harvest as much or more carbon from the atmosphere than all the terrestrial plants.
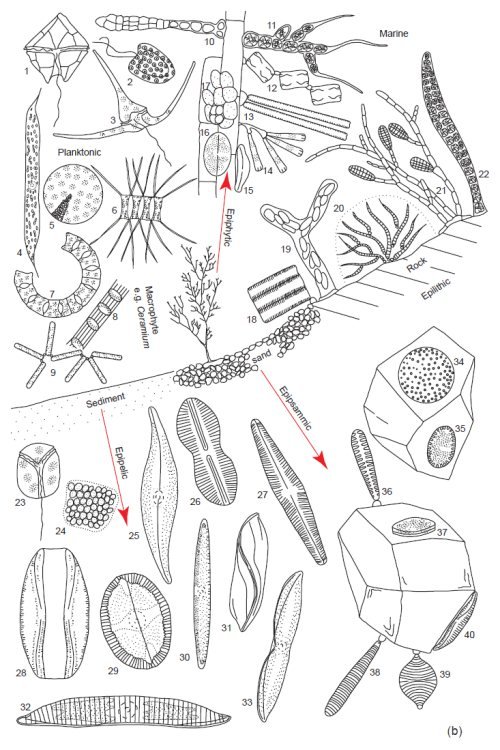
Fig. 4b. Examples of the tremendous diversity in cell form of the microscopic planktonic algae in seawater. Epilithic = growing on rocks, Epipsammic = growing in sand, Epipelic = growing in sediments, Epiphytic = growing on plants. (Round 1981). Used with permission of Cambridge University Press.
(b) Marine. Planktonic. 1. Peridinium, 2. Hymenomonas, 3. Ceratium, 4. Rhizosolenia, 5. Coscinodiscus, 6. Chaetoceros, 7. Eucampia, 8. Sceletonema, 9. Thalassionema. Epiphytic: 10. Erythrotrichia, 11. Arcrochaetium, 12. Grammatophora, 13. Synedra 14. Licmophora, 15. Amphora, 16. Cocconeis, 17. Pleurocapsa. Epilithic: 18. Fragilaria, 19. Navicula, 20. Rivularia, 21. Ectocarpus, 22. Young Bangia. Epipelic: 23. Amphidinium, 24 Holopedium, 25. Gryosigma, 26. Diploneis, 27. Navicula, 28. Amphora, 29. Mastogloia, 30. Nitzschia, 31. Amphiprora, 32. Hantzschia, 33. Tropidoneis, 34. Coscinodiscus, 35. Cocconeis, 36. Opephora, 37. Cocconeis, 38. Syndedra, 39. Raphoneis, 40. Amphora.
Non-photosynthetic protists
This is a difficult group to account for and there is much disagreement about classification. Some of the representatives of this group are the protozoa, diplomonads, parabasalids, kinetoplastids, and water molds. It is estimated that there are over 42,000 species of non-photosynthetic protists with estimates of greater than 400,000 total species in the biosphere (Chapman 2005). Found in this group also are the organelle-deficient eukaryotes, for example, the amitochondriates (lacking mitochondria). These cells have traditionally been viewed as representatives of the first eukaryotic cells, however, recently, evolutionary biologists have admitted that no truly amitochondriate eukaryotic cells have been described (Miklos Müller pers. comm.). These microbes most likely play roles in nutrient cycling and nutrient provision, but their function is not as well known as the other microbe groups.
Mosaics and metagenomes
Kurt Wise has proposed that chimeric animals like the platypus or red panda are designed as mosaics (Wise 2008). These creatures which possess an unusual eclectic mixture of phenotypic structures often populate rare ecosystems and are difficult to categorize taxonomically. In a similar fashion, we find microbes which possess a blending of characteristics or cell types; these organisms are also difficult to categorize taxonomically.
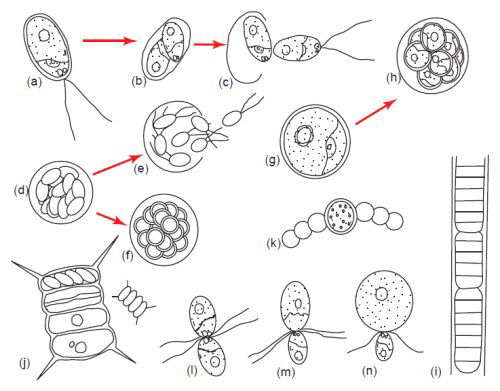
Fig. 5. Algae display tremendous diversity in cellular reproduction. (a–c) Bipartition or binary fission. (d) and (e) Zoospore formation. (f) Aplanospore formation, (g) and (h) Autospore formation. (i) Fragmentation or hormogonium formation. (j) Autocolony formation. (k) Akinete formation. (l) Isogamy. (m) Anisogamy. (n) Oogamy. Aplanospore formation is similar to zoospore formation except the spores do not have flagella but have the potential to make them. Autospores are spores which do not have the ability to make flagella. Somatic cells have the ability to become asexual spores and form extra cell wall layers as in akinete formation (Bold and Wynn 1985). Used with permission of Prentice Hall.
For example, the euglenoids are mixotrophic flagellated protists which can live a heterotropic lifestyle by engulfing food particles but they also display an autotrophic or plant-like photosynthetic lifestyle. Similarly, Hatena, a heterotrophic flagellate has been observed in association with a symbiotic algae partner which provides photosynthetic based nutrition and also an eyespot for phototaxis (Okamoto and Inouye 2005). In another example, the pathogenic eukaryotic microbe Giardia lamblia possess chimericlike genetic components which show sequence identity with both prokaryotes and eukaryotes, and they also contain a novel mitochondrium called a mitosome. Similarly, the archaea as a group possess genetic components which show identity with both prokaryotes and eukaryotes.
As noted in the Hatena example, many microbes express attributes of mosaics because they associate with a symbiotic partner which can dramatically influence the morphology, genetic makeup and physiology of the chimeric individual. This is demonstrated most dramatically in the lichen group which is clearly mosaic in appearance and in mammals which are not overtly mosaic in appearance.
Lichens
Lichens are resilient plant-like creatures composed of fungi, algae, and sometimes a bacterial partner (Armitage and Howe 2007). The interaction between the fungus and algae is intimate and involves invasion of the algae by the fungus. Unique plant-like (stem and leaf shapes) morphologies are created by the partnership which are not observed in the algae or fungus when they are separated from one another. Consequently, there is a dizzying array of lichen morphologies; it is estimated that there are 17,000 documented lichen species with an estimated 25,000 living in the biosphere (Chapman 2005). In addition roughly 20% of all fungi are lichenized and although the symbiotic partners can be cultivated separately in the lab, they do not live apart in their natural setting (Purvis 2007).

Fig. 6. A three-toed sloth Bradypus tridactylus with green algae growing on its coat. A closer examination shows that the individual hair cells are designed to accommodate the algae. This not only helps hide the sloth but provides a rich environment for other creatures. One individual sloth was found to harbor 978 beetles in its algal-laden coat (Unterthiner 2008).
Lichens are hearty creatures more resistant to extreme natural conditions than algae and fungi and consequently are found thriving in desolate environments like the surface of rocks and trees where they receive few nutrients. They can survive in extreme environments like the desert and frozen tundra and can go without water for months. They are the only known microbes to survive in the extremes of outer space. In an experiment in 2005, two different species of lichen Rhizocarpon geographicum and Xanthoria elegans were exposed to outer space on the surface of a spaceship for over 14 days. During this time the lichens were bombarded with intense solar UV light, cosmic radiation, the vacuum of outer space and wide fluctuations of temperature. Remarkably they survived the trip, and in fact their viability and ability to perform photosynthesis were unchanged when inspected in the laboratory (Sancho et al. 2007). Even though lichens can survive the extremes of outer space, they are very sensitive to environmental pollutants and do not survive well in urban areas. Because of this, they are used as “canary” species to determine the level of air pollutants in rural areas (Riddell, Nash, and Padgett 2008).
The existence of lichens raises a lot of questions regarding how we determine what a microbial species or baramin is. If lichens are individual species or baramins how do we view their genome, as a chimeric diploid or a metagenome?5 As we understand more about the contribution of microbial symbionts, the concept of the metagenome is becoming harder to ignore especially in the face of the accumulating data which shows a complex interplay between the human body and its cache of microbial symbionts.
Similar to the other microbe classes explored so far, lichens appear also to be sources of nutrition for plants and animals. Because of their ability to grow on barren landscapes lichens provide food for animals in extreme environments like the tundra. Lichens can also enhance the mineral content of ecosystems by promoting the release of minerals from rocks and by falling to the forest floor from tree or rock substrata, enriching the nutrient content of soils.
Lichens are one of several examples of macrostructures formed by the interaction of multiple microbial partners in communities. Microbes also interact intimately with non-living matter to form mosaic macro-structures which are part living and part organic or inorganic substrate. Probably the best example of this are the stromatolites which form large rock-like structures in coastal regions.
The intestinal microbiome
Humans and mammals in general are not thought of as being mosaics, yet they are inundated with symbiotic or commensual microbial flora. The most intense interactions most likely take place in the digestive tract where intestinal microflora interface in specific ways with the human tissues, influencing physiology and playing a vital role in human health. Intestinal microbes not only produce vitamins and other nutrients but they can strongly influence major body systems like the digestive, immune and nervous system. For instance microbes play an important role in determination of blood type, body weight and recent evidence suggests they can even influence brain chemistry, most likely via bacterial factors which enter the body via the circulatory system (Humphries 2009; Martin et al. 2009). The participation of microbes and microbial factors in human physiology is so significant that a recent study has proposed “that major mammalian metabolic processes are under symbiotic homeostatic control” (Martin et al. 2009). This essentially means that microbes can participate in the regulation of human body systems. Furthermore, the contribution of genes from the entire set of human symbiotic microbes increases the gene capacity of the human genome by a factor of 1000, creating a metagenome of over 20 million genes (Sleator, Shortall, and Hill 2008).
Final Observations and Closing Comments
This paper represents a first approach towards describing cellular microbes in the context of creation biology and is a call for creation microbiologists to develop this aspect of the creation model. Clearly there is much work to do just in the areas of identification and classification of microbes. For instance, as mentioned above, how do we view microbes in the context of baraminology? Perhaps one place to start is to consider why microbes were created. We know that many organisms that display a vegetative lifestyle similar to microbes were created for food (Genesis 1 and 2). Relatedly, one of the common threads that we find among the microbe groups is that they act as an organosubstrate providing nutrients for living creatures and they are involved in this process at the macro-ecosystem biogeochemical-cycling (organosubstrate) level and the micro-ecosystemorganismal level. Microbes interact in very specific ways with metazoans and thus we propose, as a first attempt at classification,6 dividing all microbes into two groups: 1) the free living organosubstrate microbes and, 2) microbes associated with metazoan organisms (Wise pers. comm.). The organism-associated bacteria would not be considered as independent free living microbes but instead as extra-corporeal or extra-cellular organelles and tissues. Thus, humans would not be considered as mosaic creations but as individuals created in the image of God who contain both prokaryotic and eukaryotic cells and tissues.
The creation-concept-derived idea that a large group of microbes were designed to associate symbiotically with organisms will also inform our understanding of disease processes and pathogenesis. For instance, creationists have predicted that pathogenic states are due to an imbalance in resident microbes or due to the presence of misplaced or altered microbes (Francis 2009; Purdom 2009; Wood 2007).
Considering the diversity and abundance of microbial groups as outlined in this paper, it is also interesting to note that the bacteria and fungi groups, which display few unique morphologies at the cell level are perhaps the most diverse microbes at the genetic level. These creatures can also be very morphogenic, forming multicellular structures, whereas the protist groups tend to be less genetically and morphogenically pliable but exhibit high extant morphological complexity and diversity. The existence of both morphological stasis and morphogenic pliability in microbes would be expected because they populate both static and changing ecosystems.
We also noted a wide ranges in sizes of microbes and it is interesting to note that bacteria are the smallest microbe at the cell level (viruses are smaller but they are not considered to be true cells) and there is a wide gulf between the size of bacteria and other microbes at the cell and genetics level.7 And as noted above, bacterial genomes remain small despite lateral gene transfer. Why are bacteria designed to remain small? Bacteria live on a lot of different creatures to a greater extent perhaps than any other cellular microbe. If one of their roles is to associate with eukaryotic cells and for example, deliver nutrition at the cell level, then it is reasonable that they would be designed to remain small so that they could access cells in tissues.
In conclusion, the recent use of molecular sequencing techniques has exposed tremendous diversity among microbe populations, and thus it is obvious that modern microbiology has just begun to describe the intricacies of the microbial world. Creationists have a unique opportunity to be at the forefront of this discipline. Classification of microbes within the creation model is an immediate problem to be solved but only one among many projects to be considered. We believe that the next most important frontier in creation microbiology will be bioremediation. In this context microbes could be used to restore disturbed ecosystems. For instance, because microbes are biogeochemical cyclers and recyclers, they could be used to reclaim polluted terrestrial, aquatic and atmospheric environments. For example, they could be used to recapture excess atmospheric carbon. And because some microbes are designed to interact with the human body, they could be used for medical bioremediation. In fact in a promising new area of medical microbiology creation microbiologists have outlined strategies for cancer biotherapy treatment using microbes (Kim 2008). Clearly there is much exciting work to be done.
References
Aiello, A. 1985. Sloth hair: Unanswered questions. In The evolution and ecology of armadillos, sloths, and vermilinguas, ed. G. G. Montgomery. Washington: Smithsonian Institute.
Anderson, K. L. 1980. Cellular origins and the three “primary kingdoms”: A critique. Creation Research Society Quarterly 16:197–202.
Armitage, M. H. and G. F. Howe. 2007. The ultrastructure of lichen cells supports creation not macroevolution. Creation Research Society Quarterly 44:107–118.
Batra, S. W. T. and L. R. Batra. 1967. The fungus garden of insects. Scientific American 217:112–120.
Baxter, J. W. and J. Dighton. 2005. Diversity-functioning relationships in ectomycorrhizal fungal communities. In The fungal community: Its organization and role in the ecosystem, eds. J. Dighton, J. White, and P. Oudemans. Boca Raton, Florida: Taylor and Francis.
Bergman, J. 1998. The unbridgeable chasm between prokaryotes and eukaryotes. In Proceedings of the Fourth International Conference on Creationism, ed. R. E. Walsh, pp. 67–77. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Bidartondo, M. I., A. M. Kretzer, E. M. Pine, and T. D. Bruns. 2000. High root concentration and uneven ectomycorrhizal diversity near Sarcodes Sanguinea (Ericacieae): A cheater that stimulates its victims? American Journal of Botany 87:1783.
Bold, H. C. and M. J. Wynn. 1985. Introduction to the algae. Englewood Cliffs, New Jersey: Prentice Hall.
Burnett J. 2003. Fungal populations and species. New York, New York: Oxford University Press.
Casjens, S. R. 2008. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Research in Microbiology 159, no. 5:340–348.
Chapman, A. D. 2005. Numbers of living species in Australia and the world. A report for the Department of Environment and Heritage. Retrieved on, April 6, 2009 from, http://www.environment.gov.au/biodiversity/abrs/publications/other/species-numbers/index.html
Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proceedings of the National Academy of Sciences 99:10494–10499.
Curtis, T. P. and W. T. Sloan. 2004. Prokaryotic diversity and its limits: Microbial community structure in nature and implications for microbial ecology. Current Opinion in Microbiology 7:221.
Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biology 6:e280 doi:10.1371/journal.pbio.0060280
Fauquet, C. M. and D. Fargette. 2005. International committee on taxonomy of viruses and the 3142 unassigned species. Virology Journal 2:64.
Francis, J. W. 2003. The organosubstrate of life: A creationist perspective of microbes and viruses. In Proceedings of the Fifth International Conference on Creationism, ed. R. L. Ivey, pp. 434–444. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Francis, J. W. 2008. The matrix. Answers 3(3):2.
Francis, J. W. 2009. Good designs gone bad. Answers 4(3):32.
Gregory, T. R. 2004. Macroevolution, hierarchy theory, and the C-value enigma. Paleobiology 30:179–202.
Groombridge, B. and M. D. Jenkins. 2002. World atlas of biodiversity: Earth’s living resources in the 21st century. Berkley, California: University of California Press.
Gruenke, J., J. Francis, and T. C. Wood. 2004. A proposal for a creationist survey of viruses. Occasional Papers—BSG: A Creation Biology Study Group 4:14.
Hawksworth, D. L. and G. M. Mueller. 2005. Fungal communities: Their diversity and distribution. In The fungal community: Its organization and role in the ecosystem, eds. J. Dighton, J. White, and P. Oudemans. Boca Raton, Florida: Taylor and Francis.
Hennigan, T. 2008. A solar powered animal? Creation Matters 13, no. 6:5.
Jaynes, R. A. and J. E. Elliston. 1980. Pathogenicity and canker control by mixtures of hypovirulent strains of Endothia parasitica in American chestnut. Phytopathology 70:453–456.
Jensen, M. A., S. M. Faruque, J. J. Mekalanos, and B. R. Levin. 2006. Modeling the role of bacteriophage in the control of cholera outbreaks. Proceedings of the National Academy of Sciences 103:4652.
Humphries, C. 2009. The body politic: The deep symbiosis between bacteria and their human hosts is forcing scientists to ask: Are we organisms or living systems. Seed 21:55.
Kim, L. 2008. Bacterial attenuation and its link to innate oncolytic potential. Answers Research Journal 1:117.
Kruetzer, A. M., M. I. Bidartondo, L. C. Grushiba, J. W. Spatafora, T. M. Szaro, and T. D. Bruns. 2000. Regional specialization of Sarcodes sanguinea on a single fungal symbiont from the Rhizopogon ellenae species complex. American Journal of Botany 87:1778.
Lane, N. 2007. Mitochondria: Key to complexity. In Origin of mitochondria and hydrogenosomes, eds. W. Martin and M. Muller. Berlin, Germany: Springer Verlag.
La Scola, B., C. Desnues, I. Pagnier, C. Robert, L. Barrassi, F. Ghislain, M. Merchat, M. Suzan-Monti, P. Forterre, E. Koonin, and D. Raoult. 2008. The virophage as a unique parasite of the giant mimivirus. Nature 455:100.
Leidy, J. 1849. On the existence of entophyta in healthy animals, as a natural condition. Proceedings of the Academy of Natural Sciences of Philadelphia 4:225–233.
Lindell, D., M. B. Sullivan, Z. I. Johnson, A. C. Tolonen, F. Rohwer, and S. W. Chisholm. 2004. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proceedings of the National Academy of Sciences 101: 11013–11018.
Margulis, M., J. Z. Jorgensen, S. Dolan, R. Kolchinsky, F. A. Rainey, and S. C. Lo. 1998. The Arthromitus stage of Bacillus cereus: Intestinal symbionts of animals. Proceedings of the National Academy of Sciences 95, no. 3:1236.
Martin, F. P., N. Sprenger, I. K. Yap, Y. Wang, R. Bibiloni, F. Rochat, S. Rezzi, C. Cherbut, S. Kochhar, J. C. Lindon, E. Holmes, and J. K. Nicholson. 2009 Panorganismal gut microbiome-host metabolic crosstalk. Journal of Proteome Research 8:2090.
Massana, R. and C. Pedros-Alio. 2008. Unveiling new microbial eukaryotes in the surface ocean. Current Opinion in Microbiology 11:213.
Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 18:405.
Okamoto, N. and I. Inouye. 2005. A secondary symbiosis in progress. Science 310:287.
Parfrey, L. W., E. Barbero, E. Lasser, M. Dunthorn, D. Bhattacharya, D. J. Patterson, and L. A. Katz. 2006. Evaluating support for the current classification of eukaryotic diversity. PLoS Genetics 12:e220.
Purdom, G. 2009. The role of genomic islands, mutations, and displacement in the origin of bacterial pathogenicity. Answers Research Journal, in press.
Purvis, O. W. 2000. Lichens. Natural History Museum, London/ Smithsonian Institution, Washington D.C. Reprinted 2007 in U. K.
Ricklefs, R. E. 2008. The economy of nature. New York, New York: Freeman.
Riddell, J., T. H. Nash, and P. Padgett. 2008. The effect of HNO3 gas on the lichen Ramalia menziessii. Flora 203:47.
Rohwer, F. and R. Edwards. 2002. The phage proteomic tree: A genome based taxonomy system for phage. Journal of Bacteriology 184:4529.
Round, F. E. 1981. The ecology of algae. Cambridge, United Kingdom: Cambridge University Press.
Sancho, L. G., R. de la Torre, G. Horneck, C. Ascaso, A. de Los Rios, A. Pintado, J. Wierzchos, and M. Schuster. 2007. Lichens survive in space: Results from the 2005 LICHENS Experiment. Astrobiology 7:443.
Sanders, I. R. 1999. No sex please, we’re fungi. Nature 399:737.
Silliman, B. R. and S. Y. Newell. 2003. Fungal farming by a snail. Proceedings of the National Academy of Sciences 100:15643-15648.
Simard, S. W., D. A. Perry, M. D. Jones, D. D. Myrold, D. M. Durall, and R. Molina. 1997. Net transfer of carbon between ectomycorrhizal tree species in the field. Nature 388:579–582.
Sleator, R. D., C. Shortall, and C. Hill. 2008. Metagenomics. Letters in Applied Microbiology 47:361.
Smith, M. L., J. N. Bruhn, and J. B. Anderson. 1992. The fungus Armillaria bulbosa is among the largest and oldest living organisms. Nature 356: 428–431. doi:10.1038/356428a0
Unterthiner, S. 2008. Three-toed sloths. BBC Knowledge Magazine 2:50.
Van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wieken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69.
Villareal, L. P. 1997. On viruses, sex and motherhood. Journal of Virology 71:859.
Whitfield, J. 2007. Underground networking. Nature 449:136.
Wilson, W. H., J. L. Van Etten, and M. J. Allen. 2008. The Phycodnaviridae: The story of how tiny giants rule the world. Current Topics in Microbiology and Immunology 328:1.
Wise, K. 2008. Mystifying mosaics. Answers 3(3):56.
Wood, T. C. 2002. The AGEing process: Rapid post-Flood intrabaraminic diversification caused by Altruistic Genetic Elements (AGEs). Origins 54:5.
Wood, T. C. 2003. Perspectives on AGEing: a young-earth creation diversification model. In Proceedings of the Fifth International Conference on Creationism, ed. R. L. Ivey, pp. 479–489. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Wood, T. C. 2007. The origin of pathogenic bacteria by degeneration or design. Occasional Papers of the BSG 10:8–9