Research conducted by Answers in Genesis staff scientists or sponsored by Answers in Genesis is funded solely by supporters’ donations.
Abstract
Recent human history represents a novel arena in which to comparatively test evolution and young-earth creation (YEC) against each other. Though both models generally accept the same sequence of events, they differ in their predictions on the relative timing of these events. One such sequence of events is the post-Columbian population collapse and recovery in Amerindian populations. I show that, when Y chromosome-based reconstructions of changes in population size are performed under the parameters of the YEC model, these reconstructions capture known post-Columbian history. This successful confirmation of the most recent, known 500 years of history permits the revisiting of the mainstream account of pre-Columbian history. Specifically, I show that modern Amerindian Y chromosome lineages descend from a group of Central Asian migrants who arrived in the Americas in the A.D. era. In combination with Amerindian archaeological history that extends into the early B.C. era, these data implied that at least one major population replacement occurred in the Americas before the arrival of Europeans. Comparison of these Y chromosome-based results with a previously discredited Amerindian origins and migration account suggests that the Amerindian account may, in fact, be authentic and that the Amerindians may have recorded the population replacement event before Y chromosome sequencing uncovered it.
Keywords: Evolution, young-earth creation, origins, predictions, Y chromosome, clock, mutation rate, population genetics, Amerindian, indigenous people, pre-Columbian, post-Columbian, replacement, Wallam Olum, Delaware
Introduction
The most recent 3,000 years of human history represent an unusual arena to test evolution against young-earth creation (YEC). After all, both sides generally agree on the sequence of events, and the short length of time—just three millennia—fits comfortably within both views. However, the relative timing of these events within each model leads to clear and contrasting predictions for human history under each model. Consequently, the fulfillment of these predictions can lead to revisions in our understanding of the history of civilization.
The field of genetics permits the analysis of these contrasting predictions. Specifically, genetics records changes in population size, as well as the contacts and separations between peoples. Evolution and YEC make different predictions about the relative genetic timing of these types of events for the most recent three millennia.
For example, evolutionists put the beginning of the history of modern Homo sapiens around 250,000 years ago (Karmin et al. 2015), and they reconstruct human history from genetics within this timeframe. Consequently, they expect the last 3,000 years of human history to show up only in the last ~1% of their historical reconstructions. In contrast, when YE creationists reconstruct human history from genetics, they stretch it out over only 4,500 years of post-Flood history (Hardy and Carter 2014; Jeanson 2019). Thus, the most recent 3,000 years represent about two-thirds of this post-Flood history, and YE creationists expect these three millennia to show up in all but the earliest third of their historical reconstructions.
Recent analyses of global Y chromosome data have confirmed the YEC expectations on a global scale (Jeanson 2019). These results have further implied that YEC expectations will be borne out at regional and local scales.
One such region is the Americas. For the pre-Columbian era, ongoing archaeological field work in combination with new technology continues to write and rewrite the history of changes in the population sizes of the Amerindians (e.g., see Canuto et al. 2018; see also de Souza et al. 2018). Furthermore, the population size in the Americas on the eve of the arrival of Columbus remains a hotly contested topic (Denevan 1992; Mann 2005). However, with respect to post-Columbian population history, the details are less disputed (McEvedy and Jones 1978; Sturtevant 1978-2004; Denevan 1992; Mann 2005). Researchers generally agree that the arrival of Columbus triggered a 300- to 400-year population decline. They also agree that this was due to enslavement, slaughter, and introduction of new diseases to which the Amerindians were not resistant. After this decline, and depending on the specific location within the Americas (e.g., Sturtevant 1978–2004; consider also the history of the Indian wars in the American West, e.g., Cozzens 2016), the Amerindian population recovery began around the 1800s (for some groups, in the 1900s), and growth generally continued into the 1900s.
Evolutionists view this history within the context of the overall evolutionary timescale, and this context makes specific predictions about the relative timing of these recent events. Specifically, evolutionary geology-based dates for the first human archaeological remains have set the overall temporal framework, and evolutionists have stretched human Y chromosome differences across this timescale. This relationship is so tight that evolutionists use the archaeology-based dates as a “sanity check” (Poznik et al. 2016, 87 of the Supplementary Information) for their genetic conclusions.1
Consequently, evolutionists explain the origin of Amerindians with a migration event across the Bering Strait and an arrival and expansion in the Americas about 15,000 years ago (Potter et al. 2018). Though archaeology has revealed the rise and fall of many pre-Columbian American civilizations (Coe and Koontz 2013; Coe and Houston 2015; Moore 2014)—e.g., the Olmec, the Mayan, and the Aztec and Incan (the fall of the latter to being precipitated by European incursion)—evolutionists attribute the origin of all of these people groups ultimately to the original crossing of the Bering Strait.
Thus, evolutionists put the relative dates for recent Amerindian events at the very tip of their historical reconstructions. For example, the recent population rebound in the 1800s represents an event from just 100 to 200 years ago. Within the framework of 250,000 years, the events of the 1800s represent the last 0.08% of evolutionary history.
From a theoretical standpoint, detecting events in this tiny of a temporal window (i.e., after 99.92% of human history has elapsed) might seem difficult. Statistically, this small window would also seem undetectable. For example, the Y chromosome tree-based branch length variation among individuals in the Karmin et al. (2015) study was around 1.5% (see the standard deviation for the Evo root in Supplemental Table 11 in Jeanson and Holland 2019). Over a 250,000-year timescale for human origins, a standard deviation of 1.5% represents 3,750 years. Yet the population recovery in the Americas happened within the last 200 years. Surely this recent event would get lost in the statistical noise associated with evolutionary analyses of the Y chromosome.
Practically, this concerns have been borne out. For example, while prior evolutionary studies have successfully detected a population decline, they have not detected the 1800s recovery. With respect to the decline, Poznik et al. (2016) showed that over 75% of Latin American men did not belong to haplogroup Q (i.e., the traditional Amerindian haplogroup) but, rather, to obviously African or European Y chromosome lineages (see Supplementary Tables 8 and 9 of Poznik et al. (2016)). In addition, Karmin et al. (2015) inferred population decline in the Amerindian population (see the Y chromosome “Andes” graph in their Figure S4A). However, neither study detected the population recovery in the 1800s.
In contrast, the YEC model makes very different predictions for the recent history of changes in the population sizes of the Amerindians. Since YEC history is only a few thousand years long, the recent population rebound in the 1800s represents an event which covers a higher percentage of total history than under the evolutionary model. Instead of representing just 0.08% of the total history, these events represent closer to 4% of the total.
Under the YEC model, theoretical and statistical considerations predict that the recovery in the 1800s should be detectable. For example, since the Y chromosome clock appears to tick every generation (Jeanson and Holland 2019), it offers, in theory, single-generation resolution. In addition, for branch lengths based on the Alpha root (see Supplemental Table 11 in Jeanson and Holland 2019), the standard deviation ranges from 1.4% to 4.2%. Over a 4,500- year post-Flood timescale, a 4.2% uncertainty represents 189 years; a 1.4% uncertainty, just 63 years. Thus, if the YEC model is correct, and if sampling of individuals is balanced and robust (i.e., see results and discussion of sampling in Jeanson 2019), then YEC timeline-based reconstruction of Amerindian history should be able to replicate the known post-Columbian history of the Americas, from the population decline through the recovery in the 1800s.
Using methods developed previously (Jeanson 2019), I attempted to reconstruct Amerindian history within the YEC framework to test whether I could detect the known post-1492 changes in Amerindian population size.
The results from these YEC-based tests prompted me to revisit the synthesis of pre-Columbian archaeology and genetics. In addition, these results prompted me to revisit some of the pre-Columbian history purportedly recorded by the Amerindians.
One account in particular, The Red Record: The Wallam Olum (McCutchen 1993) of the Lenni Lenape (Delaware) Indians, has been the focus of intense controversy. Having come to Western attention through the work of Constantine Rafinesque, this account of the Delaware origins and migration to North America has been treated by some as authentic history (McCutchen 1993). If authentic, it could reveal novel insights into the history of the Americas before European arrival. However, in 1995 a PhD thesis was published arguing “that the Walam Olum is indeed a hoax and that Rafinesque, the alleged discoverer, was actually the indisputable forger” (Oestreicher, ii–iii). I used my Y chromosome reconstructions to compare my inferred pre-Columbian history to the history described in the Wallam Olum, in order to evaluate its reliability.
Materials and Methods
Reconstruction of Amerindian Population History
In mainstream science, Y chromosome haplogroup Q is treated as the lineage of the indigenous Americans. From Supplemental Tables 3–5 of Jeanson (2019), I extracted the Hi and Lo branching dates for Amerindian individuals in haplogroup Q. Because Eskimos are mobile across the Arctic, and because some Eskimo populations still reside in Asia, I excluded them from my analyses. However, I retained those individuals in haplogroup Q from the Cachi, Wichi, and Colla populations—who are the only non-Eskimo Amerindian populations present in this particular dataset.
I identified the split date for the peopling of the Americas as the point at which a permanent break between Asians and Amerindians/Eskimos occurred in the Y chromosome tree. Effectively, I extracted the dates for node 90 and nodes 92 through 101, sorted them by date from oldest to most recent, and plotted the resultant curve (Supplemental Table 1).
To confirm that the results I observed were not an artifact of tree-building methods employed by Karmin et al. (2015), I repeated this analysis with the dates based on the previously published neighbor-joining tree based on the Alpha root (Jeanson 2019; Jeanson and Holland 2019). Effectively, I extracted the dates for nodes 421, 422, 486, 497, 501, 504, 505, and 507 from Supplemental Table 8 of Jeanson (2019); sorted them by date from oldest to most recent; and plotted the resultant curve (see Supplemental Table 2 of this paper for details).
Because the Amerindians present in haplogroup Q in Karmin et al. (2015) were only from northwest Argentina, I expanded this branch count analysis to another study. Pinotti et al. (2019) reported a Y chromosome tree for haplogroup Q individuals from up and down the Americas. Their tree included both newly sequenced individuals as part of their study, as well as previously published sequences. Though they specifically chose deep-rooting individuals to sequence, their 20 new sequences were a minority compared to the 65 previously published sequences (if we include only non-ancient DNA individuals, this number is 49, not 65—but 49 still represents the majority of the sequences).
I noticed that the evolutionary date (13,250 to 16,970 years ago) for the M3 node in haplogroup Q in Pinotti et al. (2019) overlapped the evolutionary date (14,390 to 16,480 years ago) for the M3 node in Karmin et al. (2015) (see Table S7 in for specifics; the M3 node is also labeled node 91 in Figure S3 in Karmin et al. (2015)). Therefore, I converted the evolutionary dates to YEC dates using the conversion factors for the Alpha root in Supplemental Table 4 of Jeanson (2019), which were originally used to convert data in the Karmin et al. (2015) dataset. See Supplement Table 3 in this study for details of the conversion. After converting the dates, I sorted them by date from oldest to most recent, and plotted the resultant curve (see Supplemental Table 3).
As per the findings of Jeanson (2019), I used the branch counting method only for living individuals; I excluded fossil DNA samples from my analysis. Furthermore, in online Data S4, the authors reported the evolutionary dates for some—but not all, in particularly the most recent—of the nodes in their trees displayed in Figure S1. I performed my analysis with only those nodes that had reported dates.
For the Pinotti et al. (2019) study, I defined the Amerindian split point from Asia as the point at which the last non-Amerindian lineage separated from the Amerindian ones. In Figure S1 of Pinotti et al. (2019), I chose node M930 and not node MPB001 as the split point.
The theoretical basis for this decision followed from the findings in Jeanson (2019). In Jeanson (2019), I showed that Y chromosome lineage coalescence was a function of changes in population size. For example, within the last ~600 years, the world population has increased by an order of magnitude (McEvedy and Jones 1978). Or, looking backward in time to around A.D. 1400, you could say that the world population has dropped by an order of magnitude. Conversely, looking backward in time from the present, many Y chromosome lineages from living men coalesce around A.D. 1400. Looking from A.D. 1400 backward even further in time, the world population does not shrink by another order of magnitude until pre-1000 B.C., a time gap of over 2,400 years. Consistent with this, Y chromosome lineages coalesce much more slowly in this part of the tree. Thus, for any population that has not undergone a recent explosive period of growth, the Y chromosome lineages for individual members of a population will likely coalesce over a wide range of dates. Consequently, a sudden split in a clean break; rather, because of overlapping dates for lineage coalescence, each resultant population will still show intermixing of lineages on the Y chromosome tree for dates before the split. However, for dates after the actual split happened, lineages between the two populations will not coalesce apart from contact between the two groups. Thus, I used the last coalescence date between Amerindian and non-Amerindian lineages as the best estimate for the split date between these groups.
Comparison to Known Amerindian Population History
I compared this population reconstruction to the known history of Amerindian population sizes in the Americas. Working backward in time from A.D. 1975, I reconstructed the known history in steps. First, I extracted population sizes for Amerindians from McEvedy and Jones (1978; see 270 and Fig. 4.7 on 280) from A.D. 1975 back to A.D. 1900. This showed general population growth from A.D. 1900 to A.D. 1975, punctuated by a downturn around A.D. 1950. Because my Y chromosome-based reconstructions of population history were taken from the survivors of this population downturn, I compared my reconstructions to the minimum historical population sizes. Thus, following the practice of Jeanson (2019), I converted this dynamic population growth curve to a minimum population growth curve (see Supplemental Table 4).
McEvedy and Jones (1978; see specific discussions by county in Part 4) and Sturtevant (1978–2004) indicated that the Amerinidians reached their post-1492 population nadirs pre-1900. Primarily, these occurred around the 1800s, but the specific period during the 1800s varied by geographical location. Hence, I modeled the population nadir as both 1800 and 1900, to reflect this diversity (see Supplemental Table 4).
For the 1492 population sizes, Denevan (1992) documents a wide range of estimates. The highest estimates suggest a massive population collapse followed the arrival of Columbus; the lowest suggest hardly any collapse. The methods of Jeanson (2019) did not necessitate taking a position on this debate. Again, because my Y chromosome-based reconstructions of population history were taken from the survivors of this population downturn, I needed to compare my reconstructions to the minimum historical population sizes. Effectively, this required that I draw a line backward from the nadir (i.e., somewhere in the 1800s) to the pre-Columbian time that represented the next lower population size (see Supplemental Table 4).
Unfortunately, because the dynamics of pre-Columbian population changes are still under investigation (again, see Canuto et al. 2018 as an example), this pre-Columbian data point remains unknown. To represent this uncertainty, I drew a solid population growth curve line backward from the 1800s to 1492, and then a dotted line backward from 1492 (see Supplemental Table 4).
Analysis of Wallam Olum
I used the translated text of the Wallam Olum in McCutchen (1993) to test whether the stated events in the Wallam Olum could be correlated with the history I inferred from my Y chromosome analysis, as well as with notable pre-Columbian palaeoclimatological events.
Of all the described events in the Wallam Olum, I focused on the most significant American ones. For example, the beginning of the Wallam Olum described what appeared to be a Creation-Fall-Flood-Ice Age sequence of events (Book 1, stanza 1 through Book 3, stanza 6; McCutchen 1993; see also Morris and Malone 2014), but these did not appear to take place in the Americas. Rather, after this sequence, the Wallam Olum seemed to describe an event that sounded like a crossing of the Bering Strait (McCutchen 1993; Book 3, stanzas 11 through 20). Then, in Books 4 and 5, the Wallam Olum recorded a long list of successive leaders—sachems. For some of these sachems, the Wallam Olum briefly described associated and notable events, some of which provided a basis for estimating calendar dates for the rule of each sachem.
The dates for several sachems were estimated by McCutchen (1993) via correlation of the Wallam Olum events with recorded events. For example, the Wallam Olum appeared to describe the initial arrival of the Delawares at the Atlantic Ocean: “Near Fulfilled was the sachem in sassafras country. All the Hunters reached the Sun’s Salt Sea; one more, the Ocean. Red Arrow was the sachem at the tidewater” (Book 5, stanzas 25–27; see McCutchen 1993, 124). Conversely, the Delaware wampum-based records put the arrival date of the Delaware at the Atlantic as A.D. 1396 (McCutchen 1993).
As another example, the Wallam Olum also described two encounters with whites. The first: “Mistaken was the sachem about what then came. For at this time from the Dawn Sea the Whites appeared” (Book 5, stanzas 39–40; see McCutchen 1993, 128). The second: “Watching closely was the sachem, looking seaward. For at that time from the north and south, the white people came. Friendly people, in great ships; who are they?” (Book 5, stanzas 58–60; see McCutchen 1993, 136). McCutchen (1993) associated the first encounter with Giovanni da Verrazano’s arrival in A.D. 1524; the second, with the European arrival around A.D. 1620.
Based on these dates, as well as events in post-Wallam Olum Delaware records, McCutchen (1993) estimated an average length of sachem rule to be 13.67 years (see pages 18–19 of McCutchen 1993).
From these dated encounters, I performed my own estimate of the length of sachem rule by extracting each sequential sachem name from the Wallam Olum (McCutchen 1993) to an Excel spreadsheet (Supplemental Table 5). I then counted the number of sachems who followed Mistaken up until the end of the Wallam Olum and then divided this number by 96 years (i.e., A.D. 1620–A.D. 1524 = 96 years). The result was around half the average length that McCutchen (1993) estimated for the wider timespan. I also took the average of these two average lengths of rule.
Using this range of lengths for sachem rule, I counted backward in my list of sachem names to the first sachem (“White Eagle”) who led the apparent crossing of the Bering Strait, in order to date this crossing (see Supplemental Table 5).
I also used this range of lengths for sachem rule to date two additional major events recorded in the Wallam Olum (Supplemental Table 5). The first appeared to be a major population split followed by intense conflict. Prior to the arrival in the Americas, in Book 3, stanza 19, the Wallam Olum described three major subgroups in the population that crossed the Bering Strait—“People . . . of the Eagle, of the Beaver, of the Wolf.” Then in Book 4, stanzas 10 through 14, a major population dispersal seemed to occur:
After him,
The sachem was
Chilili, the Snow Bird,
Who spoke of the south.That our people
Would be able
To grow and
Spread there.Southward went
Chilili;
Eastward went
The Beaver.To Akolaki, Snake Land,
Southern country,
Tall pine country,
Seashore country.To Eastern country,
Fishing country,
Mountain country,
Game herd country.
A few stanzas later (Book 4, stanza 17), the Wallam Olum included the following ominous description:
After Ayamek,
Ten sachems;
Much evil was then
South and eastward.
These ten sachems were unnamed in the Wallam Olum. However, whatever conflict they oversaw, they were followed by a sachem named “The Peaceful One.” In other words, the time of evil appeared to have been followed by a time of peace. I used the range of lengths for sachem rule to date this period of Delaware history (Supplemental Table 5).
The second major event was a taxing episode of drought (Book 4 stanzas 27 through 30a):
The next sachem was
Shriveled Man;
The next sachem was
Drought.There was no rain,
No food to gather;
Eastward they went
To where there was water.Beyond the pass,
In the herd country,
They found food
In the Great Plains.After Drought,
Exhaustion;
After him was
The Hardened One.
Using the range of lengths for sachem rule, I dated this period of drought (Supplemental Table 5), and I compared it to the paleoclimatic records of drought for the western and Great Plains regions of the US (Woodhouse and Overpeck 1998). I extracted the dates for multi-decadal droughts from Figure 10 of Woodhouse and Overpeck (1998).
McCutchen (1993) identified a gap of two stanzas in Book V of the Wallam Olum. Given a pattern of naming zero to two sachems per stanza, it was possible that my estimated dates were up to four sachems too recent. To account for this possibility, I added four units of sachem rule to the respective minimum calculated dates for each of my analyses (Supplemental Table 5).
Results
YEC-Based Clocks Successfully Capture Post-Columbian Amerindian History
In Jeanson (2019), the question of the root of the Y chromosome tree was left open among a range of possibilities—from the Epsilon root to the Gamma root, and any roots between these positions. To avoid prematurely picking a root, I reconstructed the Amerindian population history based on three representative root positions: Epsilon, Alpha, and Gamma. Reconstructions based on each of these root positions successfully depicted the hallmark of a population decline—namely, flat-lining in the population growth curve; and then they depicted the population recovery in the 1800s (fig. 1).
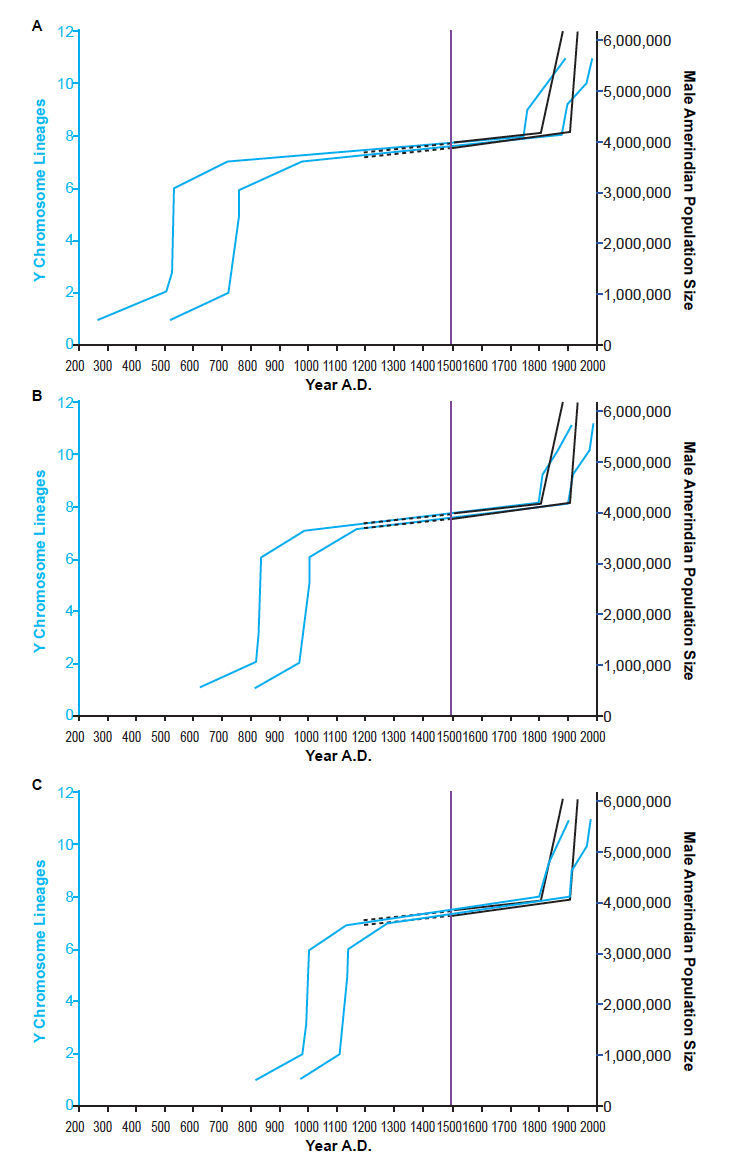
Fig. 1. Reconstruction of Amerindian population history. Using various representative root positions—i.e., Epsilon (A), Alpha (B), Gamma (C)—for the Y chromosome tree given in Karmin et al. (2015), the population history of Amerindian males was reconstructed using the branch counting method. Light blue lines represent the Y chromosome-based reconstruction. Black lines represent historical range of estimates of the minimal male population size. Solid black lines are based on more reliable historical data; dotted black lines represent the uncertainty about the pre-Columbian population sizes. The solid purple line designates the year A.D. 1492, the arrival of Columbus in the New World. The Y chromosome-based reconstructions successfully depicted both population collapse (i.e., flat-lining pre- and post- A.D. 1492) and population recovery (i.e., in the 19th century). They also suggested that the ancestors of modern Amerindians arrived in the New World in the A.D. era.
These results were not an artifact of the tree-building methods of Karmin et al. (2015). I was able to reproduce them with data based on the neighbor-joining tree in Jeanson and Holland (2019) and on the extracted data in Jeanson (2019) (see Supplemental Figure 1 in the present paper). Furthermore, these results were not an artifact of the population sampling in Karmin et al. (2015). When I reconstructed the Amerindian population history with the diversity of individuals in Pinotti et al. (2019), the shape of the population growth curve was the same, including the long flat-lining portion (fig. 2). Population recovery in the 1800s was not present, but this was likely due to the absence of reported dates for the most recent nodes depicted in their tree (e.g., see the absence of reported dates in their Data S4 for nodes CTS5173, CTS749, and Y26480 from the tree in Figure S1).
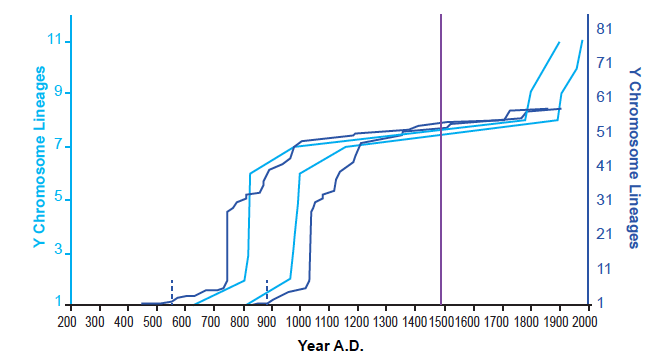
Fig. 2. Agreement on the shape of Amerindian population history between independent datasets. Using the Alpha root position for the Y chromosome tree, the population history of Amerindian males was reconstructed using the branch counting method. This was done both for the tree given in Karmin et al. (2015) and for the tree given in Pinotti et al. (2019). Light blue lines represent the reconstructions based on the Karmin et al. (2015) dataset; dark blue lines, reconstructions based on the Pinotti et al. (2019) dataset. The solid purple line designates the year A.D. 1492, the arrival of Columbus in the New World. The dotted blue lines represent my best estimate for the split date of the Amerindians from the Central Asia peoples in the Pinotti et al. (2019) dataset. Despite drawing on different population sources, both the Karmin et al. (2015)-based and Pinotti et al. (2019)-based reconstructions agreed in the overall shape of Amerindian population history.
YEC-Based Clocks Reveal Insights into Pre-Columbian History
The successful reproduction of post-Columbian history prompted me to examine the implications of my genetic analyses for pre-Columbian history. In all three reconstructions (fig. 1A–C), the arrival in the Americas occurred in the A.D. era and was quickly followed by rapid population growth and dispersion throughout North and South America.
For example, in the Epsilon root-based reconstruction, the arrival occurred somewhere between A.D. 265 and A.D. 520, and the population grew rapidly and dispersed somewhere in the range of A.D. 500 to A.D. 750 (fig. 1A; table 1). In the Alpha root-based reconstruction, the arrival occurred somewhere between A.D. 615 and A.D. 800, and the population grew rapidly and dispersed somewhere in the range of A.D. 800 to A.D. 1000 (fig. 1B; table 1). Finally, in the Gamma root-based reconstruction, the arrival occurred somewhere between A.D. 800 and A.D. 960, and the population grew rapidly and dispersed somewhere in the range of A.D. 970 to A.D. 1130 (fig. 1C; table 1).
| Y Chromosome Root | Range of dates for arrival in the Americas | Range of dates for rapid population growth and dispersal |
|---|---|---|
| Epsilon | A.D. 265 to A.D. 520 | A.D. 500 to A.D. 750 |
| Alpha | A.D. 615 to A.D. 800 | A.D. 800 to A.D. 1000 |
| Gamma | A.D. 800 to A.D. 960 | A.D. 970 to A.D. 1130 |
Since Mayan archaeology extends into the B.C. era and Olmec archaeology deeply into the B.C. era, the combination of my genetics-based sequence of events with the archaeology-based sequence of events suggested that the current Amerindian male population replaced other pre-Columbian populations. Intriguingly, the period of rapid population growth in the Epsilon root-based curve captures the time period during the collapse at Teotihuacan (Coe and Koontz 2013). Also, the period of rapid population growth in the Alpha root-based curve captures the time period during the collapse in the Mayan civilization (Coe and Houston 2015). These correlations suggested potential cause-effect relationships.
These findings made testable predictions by which they could be further evaluated and refined. Depending on the degree and level of population replacement, these results suggested that Native American lineages more ancient than Y chromosome haplogroup Q might still persist in the Americas. Given the high levels of haplogroup Q still present in Mayan populations (Perez-Benedico et al. 2016; Söchtig et al. 2015), and given the post-Columbian population collapse that occurred up and down the Americas, the more ancient lineage may have gone extinct. Nevertheless, this discovery about population replacement suggested that this ancient lineage might still exist, albeit at low levels.
These findings also predicted that additional DNA sequencing efforts from haplogroup Q Amerindians should reproduce and strengthen these population growth curve findings—provided that the sampling strategies avoid the concerns discussed in Jeanson (2019).
Did Amerindians First Document Pre-Columbian Population Replacement?
Intriguingly, I discovered that the Wallam Olum reported a sequence of events similar to the sequence implied by my Y chromosome reconstructions. First, I found agreement on the timing of the migration into the Americas. Based on the list of sachems, from the time the Delawares crossed the Bering Strait until A.D. 1620, I estimated a date of their arrival in the Americas (table 2). Using a range of estimates for the length of sachem rule, I found that all of them fell well within the dates I estimated from the Y chromosome (table 1). In fact, each estimate fell in line with the reconstructions based on each of the three Y chromosome root positions.
Second, I found agreement on the presence of other peoples in the Americas before the most recent migrants. For example, before describing the crossing of the Bering Strait, the author of the Wallam Olum described the fate and movements of enemy peoples in Asia termed “Snakes.” These Snake people left for the Americas before the Delawares did (see Book 3, stanzas 8–10). Subsequently in the Americas, the migrants repeatedly encountered and battled with Snake peoples (Book 4, stanzas 6-7, 15-16, 44; Book 5, stanzas 15–16, 42–43). Based on my Y chromosome analyses, the haplogroup Q individuals crossed the Bering Strait and wiped out other peoples who were here first—perhaps the peoples at Teotihuacan, if not the Mayan peoples. Curiously, beginning with the residents at Teotihuacan, if not with even earlier Mayan peoples, two of the major deities to receive worship in the Americas were snakes—a feathered serpent and a war serpent (Coe and Houston 2015; Coe and Koontz 2013).
Third, I found agreement on the timing of the population dispersion of the recent migrants and potential conquest of the original residents of the Americas. Based on my Y chromosome reconstructions, after the initial migration from Central Asia to the Americas, the migrant population appears to have undergone an episode of massive population growth and of population dispersion (see the sharp bend upward in the curves in figs. 1 and 2). As described above, these dispersions may have been the cause of the collapse of major civilizations in Mesoamerica. Conversely, I observed that the dates for Y chromosome-based growth and dispersion (see figs. 1 and 2 as well as discussion above) found a rough echo in the dates for the population split and conflict described above in the Wallam Olum (see tables 1 and 2). As more Y chromosome samples are obtained and sequenced, and as the split point from Central Asia is refined, this agreement between the Y chromosome-based dates and the Wallam Olum-based dates might increase.
Fourth, I found rough agreement on the size of the migrant population. For example, the author of the Wallam Olum recorded a population size of 10,000 people who crossed the Bering Strait (Book 3, stanza 18). From the Y chromosome population growth curve reconstructions, I estimated the size of the population at the time of the split from Asian peoples. Reading the y axis on the right of fig. 1(A–C) at the first data point in the growth curve revealed a population size of 500,000 or less males. Converted to a total population size, this could represent 1,000,000 people. While two orders of magnitude larger than the size depicted in the Wallam Olum, my genetics-based estimates were preliminary and based on a small sample size. As more Amerindian haplogroup Q samples are obtained, this number might drop. Either way, both the Wallam Olum and these initial Y chromosome analyses suggested that the population that crossed the Bering Strait represented a group whose size was at least an order of magnitude smaller than the estimated size of the Mayan populations in the Late Classic era (Canuto et al. 2018).
I also found correlations between events in the Wallam Olum and paleoclimatic history. The latter history depicted at least five major multi-decade episodes of drought in the pre-Columbian era. I found that at least two of these episodes overlapped with the estimated dates for the recorded drought in the Wallam Olum (i.e., compare table 2 and table 3).
| Average length of sachem rule (years) | Estimated date range for crossing the Bering Strait | Estimated era for population dispersion and major conflict | Estimated date range for drought episode |
|---|---|---|---|
| 13.67 | A.D. 280 to A.D. 335 | A.D. 335 to A.D. 554 | A.D. 663 to A.D. 745 |
| 10.53 | A.D. 588 to A.D. 630 | A.D. 630 to A.D. 799 | A.D. 883 to A.D. 946 |
| 7.38 | A.D. 896 to A.D. 926 | A.D. 926 to A.D. 1044 | A.D. 1103 to A.D. 1147 |
| Episode 1 | A.D. 250 to A.D. 300 |
| Episode 2 | A.D. 720 to A.D. 800 |
| Episode 3 | A.D. 930 to A.D. 980 |
| Episode 4 | A.D. 1130 to A.D. 180 |
| Episode 5 | A.D. 1250 to A.D. 1300 |
Discussion
Regional History Confirms the YEC Timescale and Makes Additional Testable Predictions
The regional findings of this study strengthen and underscore the global findings of Jeanson (2019). Evolutionists must now try, not only to replicate the successful capture of known Amerindian history, but also to explain why the match to the YEC expectations is so strong. Furthermore, to meet the standards articulated in this paper—and articulated over the years by evolutionists themselves, evolutionists must also publish testable predictions of their own. For example, they must publish predictions on what future studies of haplogroup Q Amerindians might reveal.
Y Chromosome Clock-Based Insights into Pre-Columbian History
The results of this study provide intriguing windows into the history of the pre-Columbian world. First, these results are consistent with large pre-Columbian population sizes. For example, in figs. 1 and 2, the flat-lining in the population growth curve extends well before A.D. 1492. If the post-Columbian population collapse was small, the flat-lining would have likely begun only shortly before A.D. 1492, and then have extended into the nineteenth century. Instead, at a minimum, the flat-lining precedes A.D. 1492 by 200 (i.e., fig. 1C) to 800 (i.e., fig. 1A) years. This is consistent with a massive drop in population numbers, in which whole villages and large regions of people—i.e., large chunks of the pre-Columbian family tree—were lost post-Columbus, but whose family tree lineages extended many years pre-Columbus, due to the sheer number of people involved.
Alternatively, these population reconstructions could be depicting extremely slow population growth prior to Columbus. However, this scenario would seem to be inconsistent with the large populations encountered in Mexico and Peru upon the arrival of the conquistadors (e.g., see Mann 2005). Furthermore, slow growth would be in contrast to the history prior to the flat-lining (i.e., rapid population growth and dispersal), and post-dating the flat-lining (i.e., 1800s and 1900s recovery); both of these two eras of history show significant rates of population growth. If the flat-lining represented slow growth, it would stand as an unusual contrast to these times of growth. More likely, the flat-lining has been caused by massive population die-off.
Second, the results of this study indicate widespread population replacement in the Americas before Columbus. However, the full extent of this replacement awaits future studies. Specifically, unbiased (i.e., no pre-screening and selection via Y chromosome typing for haplogroup Q individuals) Y chromosome sequencing of Amerindian males will be necessary to explore whether a lineage more ancient that haplogroup Q exists in the Americas, and, if it exists, at what frequency and in which populations it exists.
Could the post-Columbus population collapse have selectively wiped out this more ancient population? Might the haplogroup Q individuals have been more resistant to the causes of this population collapse? Until a more ancient lineage is found, these questions remain difficult to answer. However, the current data (figs. 1 and 2) based on the replacement (haplogroup Q) population shows evidence of post-Columbian population decline, indicating that they also were affected by the European-induced collapse. Thus, if there was a selective advantage in being part of the replacement population, it did not protect against population collapse—perhaps against extinction, but not against collapse.
Consequently, these results imply that many pre-Columbian populations have much closer genealogical relationships than mainstream science suggests. For example, given the late pre-Columbian dates for the Aztec and Incan civilizations, my Y chromosome data imply that Aztec and Incan peoples originated from the same A.D.-era source population. These Y chromosome data also imply that these Mesoamerican and South American nations had similar genealogical connections to the North American nations, such as the Navajo, Sioux, Delaware, and the like.
An additional ramification of these results touches the realm of linguistics, a field often used to explore historical relationships. Given the genealogical relationships implied by my Y chromosome results, a reevaluation of current Amerindian linguistic relationships and timelines seems warranted. Currently, the Americas have an unusual distribution of languages (Simons and Fennig 2018a, 2018b, 2018c). The Americas have an average of 12 languages per family (Simons and Fennig 2018b). Europe has 50% more—18 languages per family (Simons and Fennig 2018c). The ratio in the Pacific (41 languages per family), Asia (58 languages per family), and Africa (153 languages per family) are all much higher (Simons and Fennig 2018a, 2018b, 2018c). Furthermore, the Americas contain almost half of the world’s language families (Simons and Fennig 2018a, 2018b, 2018c). In the past, the number of Amerindian language families has been the subject of mainstream scholarly debate (e.g., see Greenberg 1987; see also Campbell 1997). Perhaps this debate should be revisited; my genetic data offer a new framework in which to do so.
New Perspective on the Authenticity of the Wallam Olum
In light of the multiple points of agreement between the genetic data in this study and the Wallam Olum, and given the palaeoclimatological agreement with the Wallam Olum, my results suggest that the Wallam Olum is an authentic account of Delaware history. If nothing else, my data suggest that Rafinesque likely did not forge the document. Surely he could not have written a fake that anticipated Y chromosome discoveries which were, at the time of the alleged forgery, still 200 years in the future. Conversely, if the Wallam Olum is an authentic history, then the agreement with my Y chromosome results suggests that my genetic findings do not represent new discoveries; rather, my genetic findings represent rediscovery of old and neglected history.
Summary and Conclusion
The successful capture of known Amerindian history underscores the utility of young-earth Y chromosome trees as a tool by which to probe the history of civilization. It also raises new challenges for the evolutionary model as it must not only replicate the success of the YEC model, but also explain why the YEC model has achieved such strong scientific confirmation. Conversely, the pre-Columbian implications of this study intimate the possibility that other novel insights into the history of civilization await more in-depth study of the Y chromosome tree.
Acknowledgements
Special thanks to Rob Carter, for his helpful interactions over a period of many months, and for sharing unpublished data that helped spur the present analysis. Many thanks as well to the Answers in Genesis librarian, Walt Stumper, whose diligent efforts in tracking down sources—even if obscure—made the present paper possible. Discussions with Steve Austin of the Medieval Warming Period were also helpful in identifying events to check in the Wallam Olum. Thanks as well to the reviewers, and to Roger Patterson among others who offered helpful feedback.
References
Campbell, Lyle. 1997. American Indian Languages: The Historical Linguistics of Native America. New York: Oxford University Press.
Canuto, Marcello A., Francisco Estrada-Belli, Thomas G. Garrison, Stephen D. Houston, Mary Jane Acuña, Milan Kováč, Damien Marken, et al. 2018. “Ancient Lowland Maya Complexity as Revealed by Airborne Laser Scanning of Northern Guatemala.” Science 361, no. 6409 (28 September: eaau0137. doi: 10.1126/science.aau0137.
Coe, Michael D., and Stephen Houston. 2015. The Maya. New York: Thames & Hudson Inc.
Coe, Michael D., and Rex Koontz. 2013. Mexico: From the Olmecs to the Aztecs. New York: Thames & Hudson Inc.
Cozzens, Peter. 2016. The Earth Is Weeping: The Epic Story of the Indian Wars for the American West. New York: Alfred A. Knopf.
de Souza, Jonas Gregorio, Denise Pahl Schaan, Mark Robinson, Antonia Damasceno Barboas, Luiz E. O. C. Aragão, Ben Hur Marimon Jr., Beatriz Schwantes, et al. 2018. “Pre- Columbian Earth-Builders Settled Along the Entire Southern Rim of the Amazon.” Nature Communications 9, no. 1125 (27 March). doi:10.1038/s41467-018-03510-7.
Denevan, William M. ed. 1992. The Native Population of the Americas in 1492. Madison, Wisconsin: The University of Wisconsin Press.
Greenberg, Joseph H. 1987. Language in the Americas. Stanford, California: Stanford University Press.
Hardy, Chris, and Robert Carter. 2014. “The Biblical Minimum and Maximum Age of the Earth.” Journal of Creation 28, no. 2 (August): 89–96.
Jeanson, Nathaniel T. 2019. “Testing the Predictions of the Young-Earth Y Chromosome Molecular Clock: Population Growth Curves Confirm the Recent Origin of Human Y Chromosome Differences” Answers Research Journal 12 (December 4): 405–423.
Jeanson, Nathaniel T., and Ashley D. Holland. 2019. “Evidence for a Human Y Chromosome Molecular Clock: Pedigree-Based Mutation Rates Suggest a 4,500-Year History for Human Paternal Inheritance” Answers Research Journal 12 (December 4): 393–404.
Karmin, Monika, Lauri Saag, Mário Vicente, Melissa A. Wilson Sayres, Mari Järve, Ulvi Gerst Talas, Siiri Rootsi, Anne-Mai Ilumäe, et al. 2015. “A Recent Bottleneck of Y Chromosome Diversity Coincides With a Global Change In Culture.” Genome Research 25, no. 4 (April 23): 459–466.
Mann, Charles C. 2005. 1491: New Revelations of the Americas Before Columbus. New York: Alfred A. Knopf.
McCutchen, David, trans. 1993. The Red Record: The Wallam Olum. Garden City Park, New York: Avery Publishing.
McEvedy, C., and R. Jones. 1978. Atlas of World Population History. Middlesex, England: Penguin Books.
Moore, Jerry D. 2014. A Prehistory of South America: Ancient Cultural Diversity on the Least Known Continent. Boulder, Colorado: University Press of Colorado.
Morris, John D. and Bruce Malone. 2014. “The Red Record.” Acts & Facts 43, no. 1 (December 30): 16–18.
Oestreicher, D.M. 1995. The Anatomy of the Walam Olum: A 19th Century Anthropological Hoax. Ph.D. dissertation, Rutgers University. New Brunswick, New Jersey.
Perez-Benedico, David, Joel La Salvia, Zhaoshu Zeng, Giselle A. Herrera, Ralph Garcia-Bertrand, and Rene J. Herrera. 2016. “Mayans: A Y Chromosome Perspective.” European Journal of Human Genetics 24 (September): 1352–1358.
Pinotti, Thomas, Anders Bergström, Maria Geppert, Matt Dawn, Dominique Ohasi, Wentao Shi, Daniela R. Lacerda, et al. 2019. “Y Chromosome Sequences Reveal a Short Beringian Standstill, Rapid Expansion, and Early Population Structure of Native American Founders.” Current Biology 29, no. 1 (7 January): 149–157.
Potter, Ben A., James F. Baichtal, Alwynne B. Beaudoin, Lars Fehren-Schmitz, C. Vance Haynes, and Vance T. Holliday 2018. “Current Evidence Allows Multiple Models For the Peopling Of the Americas.” Science Advances 4, no. 8 (1 August): eaat5473. doi: 10.1126/sciadv.aat5473.
Poznik, G. David, Yali Xue, Fernando L. Mendez, Thomas F. Willems, Andrea Massaia, Melissa A. Wilson Sayres, Qasim Ayub, et al. 2016. “Punctuated Bursts in Human Male Demography Inferred From 1,244 Worldwide Y-chromosome Sequences.” Nature Genetics 48, no. 6 (25 April): 593–599.
Simons, Gary F., and Charles D. Fennig. eds. 2018a. Ethnologue: Languages of Asia. Dallas, Texas: SIL International Publications.
Simons, Gary F., and Charles D. Fennig. eds. 2018b. Ethnologue: Languages of the Americas and the Pacific. Dallas, Texas: SIL International Publications.
Simons, Gary F., and Charles D. Fennig, eds. 2018c. Ethnologue: Languages of Africa and Europe. Dallas, Texas: SIL International Publications.
Söchtig, Jens, Vanesa Álvarez-Iglesias, Ana Mosquera-Miguel, Miguel Gelabert-Besada, Alberto Gómez-Carballa, and Antonio Salas, et al. 2015. “Genomic Insights On the Ethno- History of the Maya and the ‘Ladinos’ from Guatemala.” BMC Genomics 16, no. 1 (February 25): 131.
Sturtevant, William C. ed. 1978–2004. Handbook of North American Indians. Washington, DC: Smithsonian Institution.
Woodhouse, Connie A., and Jonathan T. Overpeck. 1998. “2000 Years of Drought Variability in the Central United States.” Bulletin of the American Meteorological Society 79, no. 12 (December): 2693–2714.
Supplemental Files
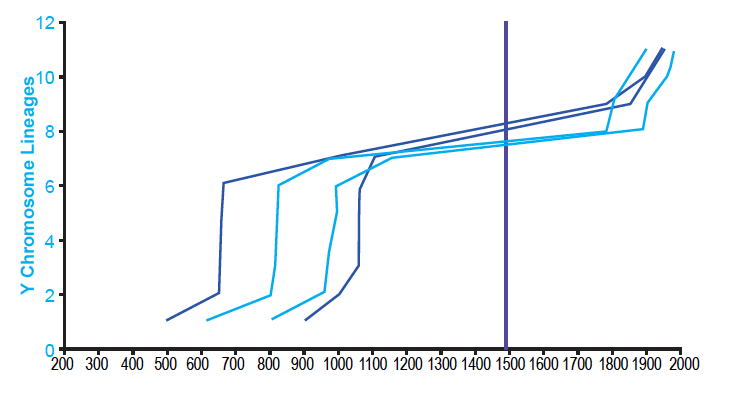
Supplemental Fig. 1. Agreement on the shape of Amerindian population history despite differing tree-building methodologies. Using the Alpha root position for the Y chromosome tree, the population history of Amerindian males was reconstructed using the branch counting method. This was done both for the data based on the tree given in Karmin et al. (2015) and for the data based on the tree in Jeanson and Holland (2019). Light blue lines represent the reconstructions based on the Karmin et al. (2015) data; dark blue lines, reconstructions based on the Jeanson and Holland (2019) data. The solid purple line designates the year A.D. 1492, the arrival of Columbus in the New World. Despite different tree-building methods, both the Karmin et al. (2015)-based and Jeanson and Holland (2019)-based reconstructions agreed in the overall shape of Amerindian population history.
Supplemental Table 1. Karmin-based data points will download when link is clicked (XLS).
Supplemental Table 2. Neighbor-joining tree-based data points will download when link is clicked (XLS).
Supplemental Table 3. Pinotti-based data points will download when link is clicked (XLS).
Supplemental Table 4. Derivation of historical population size data will download when link is clicked (XLS).
Supplemental Table 5. List of New World sachems will download when link is clicked (XLS).