The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
Despite the fact that no extant reptile species have feathers, and that the only extinct flying reptiles (the pterosaurs) had membranous wings, evolutionists assert that several dinosaur species had feathers. They go so far as to claim that birds are feathered dinosaurs. Evolutionists point to collagenous filaments in fossil remains of dinosaurs to support their theory of protofeathers in dinosaurs. But what do the genes say? Is it possible for reptiles such as dinosaurs or other groups, such as crocodilians (crocodiles, alligators, caimans, and gharials), which are considered to be archosaurs along with dinosaurs and birds, to have feathers based on genetic evidence? A study from recent years indicated that 193 keratin and non-keratin genes are associated with the formation of feathers in chicken. What is the distribution of these genes in extant reptile species? How are these genes regulated between birds and reptiles? The results show that there are many common genes regarding feather and scale formation between birds and reptiles. None of the examined reptile species have all the genes necessary for feather formation. Furthermore, if they did, these genes would also have to be regulated at the same time and place in order to transform reptile scales into avian feathers. Interestingly, turtles are the reptile group that is the most enriched in feather-associated genes, in contrast with crocodiles. Thus, feather formation does not appear to be characteristic within each of the alleged archosaurian lineages going back to dinosaurs. A high number of keratin genes in an organism does not necessarily mean they are used in feather formation. Even some fish species also have a large number of keratin genes. Also, the genetic elements of the regulatory regions of some feather-associated genes shared between birds and reptiles could be interpreted to show that some genes appear to undergo similar regulation, but other genes appear to undergo different regulation.
Keywords: feather, birds, dinosaurs, turtles, crocodiles, archosaurs, fish, BLAST, BMP2, SHH, FGF10, promoter
Introduction
A hot topic in recent years in both evolutionist and creationist circles is the question of whether some dinosaur species had feathers. Evolutionists claim that some reptiles indeed may have had feathers as forerunners to birds and that birds themselves are dinosaurs. Some creationists, on the other hand also hold to the idea of feathered dinosaurs, but only as a question of classification, rather than origin (McLain et al. 2023). The claim has been made by some that several fossil reptiles show evidence of having feathers. These include filamentous integumentary structures in patches on the animal’s skin (Xu and Guo 2009). Some creationists have contested this claim and stated that some of these collagen fibers are decayed skin fibers, and not feathers (Tay 2019).
Morphological evidence can sometimes be ambiguous, and fossil remains can be interpreted in any number of ways. However, the genotype determines the phenotype. In other words, our genes determine what we look like physically. What genes are responsible for the structural elements in feathers? Which genes are responsible for feather development and formation? How is the whole process regulated? Are there even any ‘feather’ genes in reptiles or other vertebrate groups? Answering these questions can help get closer to finding out whether dinosaurs or other reptiles indeed had feathers.
A study by Lowe et al. (2015) found 193 genes in chicken (Gallus gallus) that were associated with the formation of feathers. Of these, 60 belonged to the keratin gene family, whereas the remaining 133 genes played various other functions in feather formation. The beta-keratin gene is a member of a large family of genes coding for structures found in both birds and reptiles, such as feathers, scales, beaks, and claws (Alibardi et al. 2009). Since this gene family is so diverse and because it is so central to feather formation, these genes were treated separately from the non-keratin genes. Non-keratin genes have various functions that also take part in the development and formation of feathers, but do not belong to the keratin gene family itself.
The big question is, do these genes have homologs (genes of similar structure and function) in reptiles? If so, how many, and in what kinds of reptiles? Where should we begin our quest for feather genes in dinosaurs? It is quite remarkable that no reptile species alive today has feathers. More importantly, those reptiles that did fly, namely pterosaurs had membranous wings devoid of feathers.
Archosaurs are the alleged ancestors of pterodactyls and the dinosaurs. According to evolutionary theory, birds and crocodilians are the only living archosaurs. Dinosaurs and pterodactyls have gone extinct, and thus their genetic material is inaccessible to us. However, according to evolution, crocodiles share a relatively recent common ancestor with birds. Therefore, it would be interesting to study the distribution of feather gene homologs in different extant reptile groups, such as lizards, snakes, turtles, geckos, the tuatara, and crocodiles (with a special focus on this group due to its alleged evolutionary connection to Archosaurs).
The hypothesis tested here is this: if dinosaurs did indeed have feathers, then we should find feather development-associated genes more abundantly in those extant reptile groups which are their closest relatives. These would include crocodiles, since they together with dinosaurs are allegedly archosaurs. Furthermore, at least some modern reptiles (being the descendants of feathered dinosaurs) should have feathers, and their genes should also be regulated similarly to the way they are regulated in birds. However, if dinosaurs did not have feathers, then we would likely see some other reptile group with more feather development-associated genes, along with similar genetic regulation.
Materials and Methods
Feather gene sequences
A list of 193 Ensembl gene ids was extracted from the supplementary data from Lowe et al (2015). Of the 133 non-keratin genes, protein sequences were retrieved for 118 of them from the galGal3 version of the chicken genome. A further 11 sequences were retrieved from the protein database at NCBI (National Center for Biotechnology Information). Protein sequences for all 60 of the keratin genes were also retrieved, making 189 protein sequences in total. These 193 feather-associated genes from chicken are listed in Supplementary File 1.
Methods
A Biopython script was written to query these protein sequences against the NCBI database using the protein Basic Local Alignment Search Tool (BLASTp). Queries were performed against ‘Reptilia’ and ‘fish’. Sequence homology was declared between a query protein and a database protein if the alignment’s e-value was below 1E-10. Figures were generated using the ‘ggplot2’ within R, version 4.1.0. Venn diagrams were created using the ‘Venn diagram’ tool at https://bioinformatics.psb.ugent.be/webtools/Venn. The Kruskal-Wallis test was run in R using the ‘kruskal.test’ command.
Promoter analysis
Promoter sequences were retrieved from the NCBI BLAST website using chicken protein sequences of the genes BMP2, SHH, and FGF10 as queries in the tblastx algorithm. The 2 Kbp promoter sequence upstream of the ATG start codon was retrieved for selected species. These promoters were then analyzed using the online Multiple Em for Motif Elicitation (MEME) software (version 5.5.1) (Bailey and Elkan 1994) at https://meme-suite.org/meme/tools/meme. A maximum of seven motifs were searched for, each of which was between 6–20 bp, occurring any number of times in the promoters.
All supplementary data and figures are available on Zenodo at https://zenodo.org/record/7809453#.ZDCyNfbMLrc.
Results
Beta-keratin gene matches
In order to understand the results from this analysis, let us first briefly review the concept of homologous, orthologous, and paralogous genes from a creationist perspective. Homologous genes have the same or very similar function in two different organisms. Orthologous genes are homologous genes that are present in two species after they have split off from one another. Should such an orthologous gene be duplicated within one of the species, the copy of the original gene is a paralogous gene. Homologous genes between species can be viewed as independent design elements, rather than evolutionary relations.
Figs. 1 and 2 show the result of the BLASTp search of the 60 keratin chicken proteins against Reptilia and fish in the NCBI protein database. Fourteen species had five or more BLASTP hits. Of the 14, ten are turtle species, and the remaining four are crocodiles. The two turtle species with the most homologs are Mauremys mutica, with 57 hits (95% of all 60 keratin proteins), and Gopherus flavomarginatus, with 47 homologs (78.3%). The crocodile species Alligator mississippiensis (the American alligator) had 44 homologs (73.3%). A. mississippiensis is followed by five turtle species with a decreasing number of homologs down to 17. The beta-keratin homologs found for each reptile and fish species can be found in Supplementary Files 2 and 3, respectively.
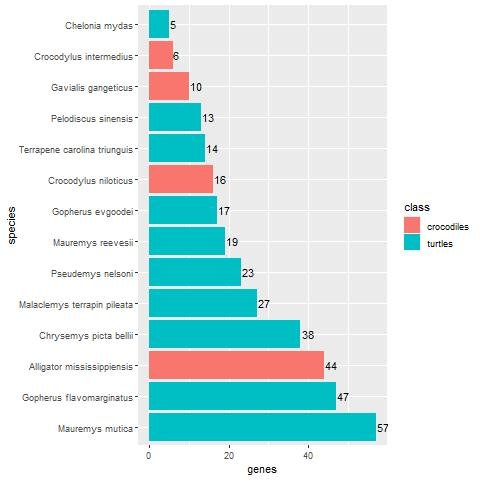
Fig. 1. Bar-plot showing the number of keratin homologs in various reptile groups, broken down per species. Crocodiles are shown in red, whereas turtles are shown in blue. In general, turtles have significantly more keratin homologs compared to crocodiles (p = 8.04E-7).
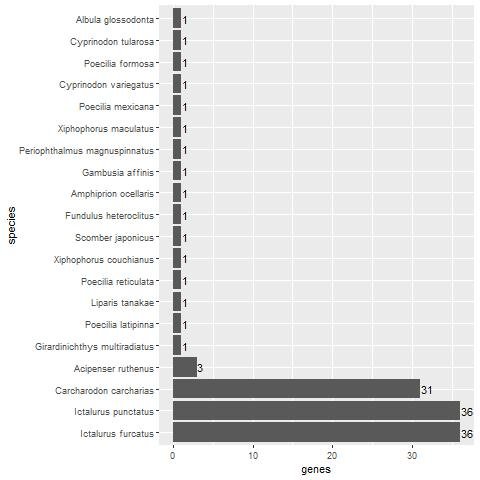
Fig. 2. Bar plot showing the number of keratin homologs in various fish species. Three species, C. carcharias, I. punctatus, and I. furcatus have a significant number of keratin homologs.
The Kruskal-Wallis test was used to determine differences in the mean number of keratin homologs per reptile group. The test returned a p-value of 8.04E-7. Thus, it is highly statistically significant that keratin homologs are the most enriched in turtles compared to all other reptile groups.
This data suggests that keratin genes are more enriched in turtles than crocodiles. Indeed, Li et al. (2013) found 200 beta-keratin genes for three turtle species, namely Chrysemys picta, Pelodiscus sinensis, and Chelonia mydas. Of these it is noteworthy that Ch. mydas is a sea turtle.
The 60 keratin genes were matched with fish genes as a negative control. Fish are not expected to have feathers due to their aquatic environment. Interestingly, three fish species also had a high number of keratin protein homologs, namely Carcharodon carcharias with 31 homologs (51.7% of the 60 chicken beta-keratin genes), and Ictalurus punctatus and Ictalurus furcatus with 33 homologs each (55%). These results strongly indicate that vertebrate animals having a high number of beta-keratin genes does not necessarily mean that they are used for making feathers. As we can see, A. mississippiensis, an alleged descendent of archosaurs along with birds, lacks more than a quarter of the keratin genes with homologs in chicken.
Non-keratin gene matches
The picture is similar when examining the 133 non-keratin protein homologs. Here 66 species have at least one homolog (see fig. 3). The five species with the highest number of homologs are turtles: Chelonia mydas, Chrysemys picta belii, Gopherus evgoodei, Chelonoidis abingdonii, and Dermochelys coriacea, with 113 (87.6%), 109 (84.5%), 108 (83.7%), 105 (81.4%), and 102 (79.1%) homologs, respectively, that matched non-keratin genes in chicken. A. mississippiensis comes in eighth place with 100 homologs (77.5%).
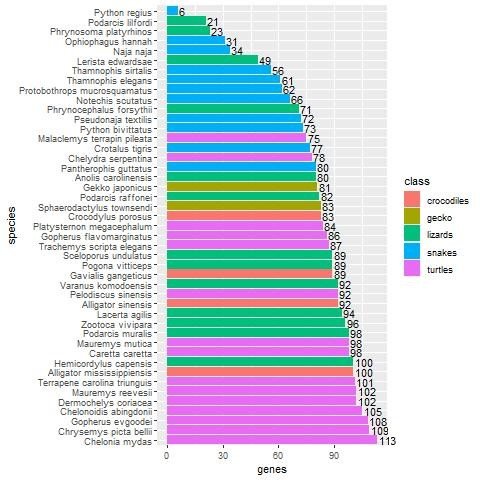
Fig. 3. Bar plot showing the number of non-keratin homologs in various reptile groups, broken down by species. Crocodiles are shown in red, gecko in dark yellow, lizards in green, snakes in blue, and turtles in purple. In general, turtles have a significantly higher number of non-keratin homologs (p = 2.2E-16).
When compared with fish, we find that Acipenser ruthenus (the Sterlet sturgeon) has almost the same number of non-keratin protein homologs as A. mississippiensis (99, see Supplementary File 3 and Supplementary fig. 1). The top 21 fish species with at least 45 non-beta-keratin homologs can be seen in fig. 4. When comparing the non-keratin homologs in the Venn diagram in fig. 5 between A. mississippiensis and A. ruthenus, we can see that they have 80 homologs in common (Jaccard Coefficient Value = 0.672). The non-beta keratin homologs found for each reptile and fish species can be found in Supplementary File 2 and 3, respectively.
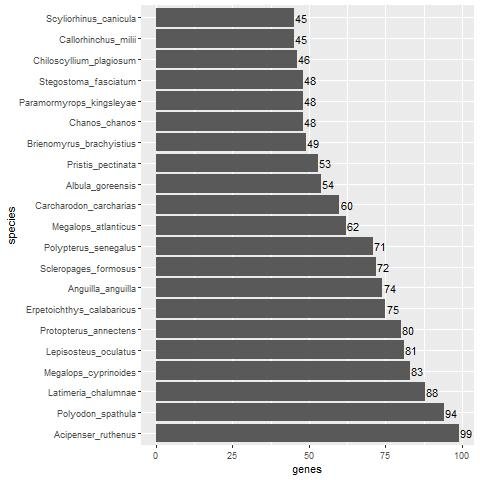
Fig. 4. Bar plot showing the number of non-keratin homologs in various fish species with more than 45 homologs.
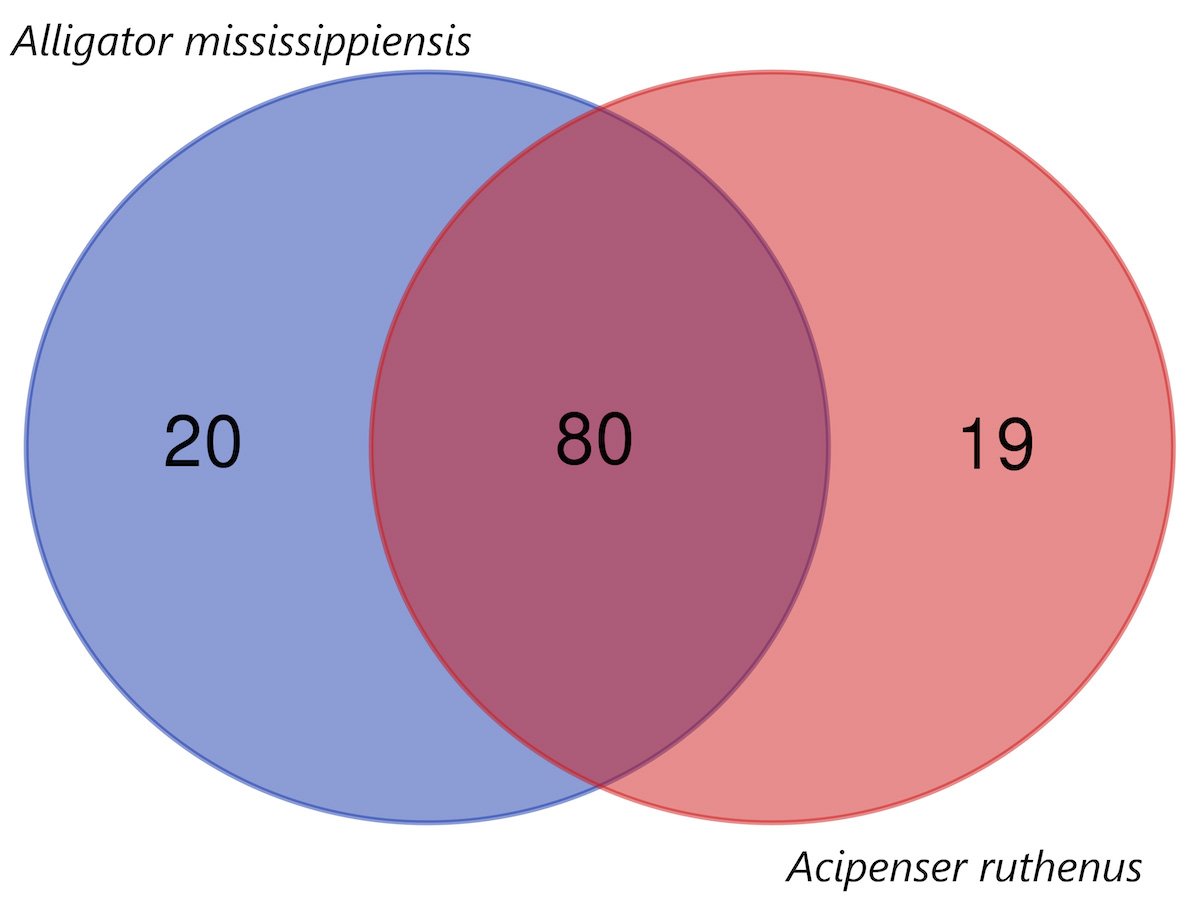
Fig. 5. Venn diagram showing the number of nonkeratin homologs between Alligator mississippiensis and Acipenser ruthenus.
Furthermore, the 133 non-keratin genes were not present in all the 66 reptile species found by the BLASTp searches. The three most common genes are TYRP1, present in 43 of the 65 reptile species (65.2%), BMP2, and MMP2 (both present in 42, or 63.6% of the species). For a list of the top ten most common genes found in reptile species, see Table 1.
| Gene | No. species | Function |
|---|---|---|
| TYRP1 | 43 | Tyrosinase related protein 1 |
| BMP2 | 42 | Bone morphogenetic protein 2 |
| MMP2 | 42 | Matrix metallopeptidase 2 |
| COL1A2 | 41 | Collagen type I, alpha 2 chain |
| HTRA1 | 41 | HtrA serine peptidase 1 |
| ADAM10 | 40 | ADAM metallopeptidase domain 10 |
| BMP4 | 40 | Bone morphogenetic protein 4 |
| EDNRB2 | 40 | Endothelin receptor B subtype 2 |
| TYR | 40 | Tyrosinase |
| WSB1 | 40 | WD repeat and SOCS box containing 1 |
The Kruskal-Wallis test was also used to determine differences in the mean number of non-keratin homologs per reptile group. The test returned a p-value of less than 2.2E-16. Thus, it is highly statistically significant that non-keratin homologs are the most enriched in turtles as opposed to all other reptiles.
Let us look at several of the key genes that take part in feather formation. The main shaft of the feather is called the rachis. Barbs branch off from the rachis on both sides, which are held together by barbules on both sides of the barbs, both anteriorly and posteriorly. Lin et al. (2013) analyzed the interactions between several genes involved in feather formation.
They found that the genes BMP4 and noggin (NOG) play a balanced, antagonistic role in the formation of feathers and the branching of barbs from the rachis. High BMP4 concentrations promotes rachis formation, whereas high levels of NOG promote rachis and barb branching. The WNT3A gene is responsible for bilateral or radial feather symmetry. FGF10 plays an important role in the formation of dermal papillae, which are the actively dividing tissues in the epidermis that give rise to the feather itself. The feather first takes on a plate-like shape, but as it develops, cells in the plate epithelium of the feather undergo apoptosis (cell death), to reveal spaces in between barbs. The spacing and pattern of the feather barbs are influenced by the expression of the BMP genes and NOG (Yu et al. 2002).
Only 10 of the 66 reptile species found in the BLASTp analyses had all three BMP genes (BMP2, BMP4, and BMP7) together with NOG, namely Alligator mississippiensis, Chrysemys picta bellii, Crotalus tigris, Gekko japonicus, Notechis scutatus, Pantherophis guttatus, Pelodiscus sinensis, Podarcis muralis, Protobothrops mucrosquamatus, and Python bivittatus. In contrast, only Pelodiscus sinensis and Gekko japonicus had these genes as well as WNT3 and FGF10. In addition to these previous genes, only Gekko japonicus contained SHH, an important gene expressed in the marginal plate of the feather (Harris, Fallon, and Prum 2002).
Promoter analysis
Promoter analysis was performed for the 2 Kbp sequence region directly upstream of the ATG start codon in five selected species for the genes BMP2, SHH, and FGF10. The results can be seen in figures 6A-C. The predicted sequence motifs in the BMP2 promoters of Gallus, Alligator, Mauremys, Chelonoidis, and Acipenser are quite dissimilar, except for a region of homology between Mauremys and Chelonoidis including the motifs CTTCCCTCYCYCCCY and YSSCGCTGCTGCTGCYVCK (fig. 6A).

Fig. 6. The result of the MEME analysis of the BMP2, SHH, and FGF10 promoters in chicken, various reptile groups, and fish. Gallus = Gallus gallus, Alligator = Alligator mississippiensis, Mauremys = Mauremys mutica, Chelonoidis = Chelonoidis abindonii, Acipenser = Acipenser ruthenus, Chrysemys = Chrysemys picta bellii, Dermoclemys = Dermoclemys coriacea, Malaclemys = Malaclemys terrapin pileate.
In contrast, the promoter regions of the gene SHH for Gallus, Alligator, Chrysemys, and Chelonoidis appear to have quite similar motif content over a larger region of the promoter (fig. 6B). This motif pattern, however, is unrecognizable in the SHH promoter of A. ruthenus.
Lastly, the FGF10 promoters in Dermochelys, Malaclemys, Chelonoidis, and Chrysemys also seem to be similar over a very large portion of the promoter (fig. 6C). However, this motif pattern cannot be discerned in the FGF10 promoter of Gallus, where the only possible motif pattern similarity is at the very front of the promoter with the RDSAGRVTAAAAGGVAAGAG motif, two TCBCAYCAGARGCWSCWKCC motifs, and a GGGRGBSAGRMCTCGRWGG motif.
Discussion
No living reptile has feathers, and the ones which flew had membranes, not feathers. The results presented here suggest that keratin genes are most abundant in turtles compared to all other reptile groups. The presence of so many keratin genes in turtles could coincide with giving structure to the ventral plastron and dorsal carapace (Li et al. 2013). Li et al. also found that 16 of the 17 beta-keratins found in Pseudoemys nelsoni were expressed in the shell skin and soft epidermis, and were also specific to turtles. The seventeenth beta-keratin gene shared between turtles and birds was expressed specifically in the claws, and not in the scales. According to Alibardi, Toni, and Valle (2007), turtles have many beta-keratins because it adds rigidity to their shells, a great adaptational advantage, and one could think, a design feature.
The fact that turtles are the reptiles with the most keratin and non-keratin genes should be of concern to evolutionists. Turtles are the least related to all the other reptiles, being anapsids (these reptiles have no fenestra between their jugal, squamosal, parietal, and postorbital skull bones). Some evolutionists place turtles as sister group to all other reptiles (Chiari et al. 2012). Others have tried to place them next to crocodiles (Cao et al. 2000), and yet others have placed them with Lepidosauria (lizards, snakes, tuatara) (Becker, Valverde, and Crother 2011).
As we have seen, many of the keratin and non-keratin genes that play a role in feather formation in the chicken are absent in reptiles. Those reptile species with the most keratin and non-keratin genes in common with birds still have scales. This also suggests that the regulation of these genes is also different than in birds. Some fish species also have a high number of non-keratin genes, yet fish are not the ancestors of birds, according to evolutionary theory.
Evolutionists might say there is no problem, because changes in the regulation of different keratin and non-keratin genes may be able to transform reptilian scales into avian feathers. However, there is no evidence for this. Computer simulations with the PromMute software have shown that appropriate-length transcription factor binding sites cannot form by random mutations, let alone in combinations (Cserhati 2012).
The promoter analysis results shown here seem to suggest that some feather development-related genes have very dissimilar promoter regions, and therefore undergo divergent gene regulation. Other genes seem to indicate similar gene regulation due to similar putative motif content. However, as in the case of FGF10, notable differences in sequence motif content between chicken and reptiles was quite apparent. The regulatory motif content of the reptile FGF10 promoters might be a turtle-specific ensemble of transcription factor binding sites. Gene regulation seems to be similar between chicken and the alligator in the SHH promoter, but different in the promoter of BMP2.
Besides having the appropriate gene repertoire for developing feathers along with their corresponding promoter regions, further genetic regulatory elements also have a say in how these genes are expressed. These elements may include enhancer elements, sometimes located on other chromosomes, as well as regulatory RNA elements, such as microRNA (miRNA), antisense RNA, short non-coding RNA (ncRNA), and long non-coding RNA (lncRNA) elements. Genetically speaking the development of feathers is not a trivial thing; rather it is a very complex molecular process involving many factors that are too numerous and intricate for evolution, based on merely random mutations to accomplish.
Summary and Conclusion
In conclusion, we can say that there are many common genes between birds and reptiles regarding the formation of feathers and scales. However, there are also crucial differences. Not all reptiles have all the genes necessary for feather formation. But, even if they did, these genes would also have to be regulated at the same time and place in order to transform scales into reptiles. Interestingly, turtles are the reptile group that is most enriched in feather-associated genes, in contrast with crocodiles, which are considered archosaurs along with birds. Thus, the alleged archosaurian lineages going back to dinosaurs do not appear to be especially geared genetically toward feather formation. When looking at the regulatory regions of feather-associated genes shared between birds and reptiles, some genes appear to undergo similar regulation, yet disparate regulation also seems to be the case between the promoters of birds and reptiles.
References
Alibardi, Lorenzo, Mattia Toni, and Luisa Dalla Valle. 2007. “Hard Cornification in Reptilian Epidermis in Comparison to Cornification in Mammalian Epidermis.” Experimental Dermatology 16, no. 12 (December): 961–976.
Alibardi, Lorenzo, Luisa Dalla Valle, Alessia Nardi, and Mattia Toni. 2009. “Evolution of Hard Proteins in the Sauropsid Integument in Relation to the Cornification of Skin Derivatives in Amniotes.” Journal of Anatomy 214, no. 4 (April): 560–586.
Bailey, Timothy L., and Charles Elkan. 1994. “Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers.” In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, 28–36. Menlo Park, California: AAAI Press.
Becker, Rosemary E., Roldán A. Valverde, and Brian I. Crother. 2011. “Proopiomelanocortin (POMC) and Testing the Phylogenetic Position of Turtles (Testudines).” Journal of Zoological Systematics and Evolutionary Research 49, no. 2 (May): 148–159.
Cao, Ying, Michael D. Sorenson, Yoshinori Kumazawa, David P. Mindell, and Masami Hasegawa. 2000. “Phylogenetic Position of Turtles Among Amniotes: Evidence From Mitochondrial and Nuclear Genes.” Gene 259, nos. 1–2 (23 December): 139–148.
Chiari, Ylenia, Vincent Cahais, Nicolas Galtier, and Frédéric Delsuc. 2012. “Phylogenomic Analyses Support the Position of Turtles as the Sister Group of Birds and Crocodiles (Archosauria).” BMC Biology 10 (27 July): 65.
Cserhati, Mátyás. 2012. “Prommute—A Promoter Mutation Simulation for Modeling the Evolution of Genetic Regulatory Elements.” Journal of Computer Science and Systems Biology 5, no. 2: 74–80.
Harris, Matthew P., John F. Fallon, and Richard O. Prum. 2002. “Shh-Bmp2 Signaling Module and the Evolutionary Origin and Diversification of Feathers.” Journal of Experimental Zoology 294, no. 2 (15 August): 160–176.
Li, Yang I., Lesheng Kong, Chris P. Ponting, and Wilfried Haerty. 2013. “Rapid Evolution of Beta-Keratin Genes Contribute to Phenotypic Differences That Distinguish Turtles and Birds from Other Reptiles.” Genome Biology and Evolution 5, no 5 (May): 923–933.
Lin, Sung-Jan, Randall B. Wideliz, Zhicao Yue, Ang Li, Xiaoshan Wu, Ting-Xin Jiang, Ping Wu, and Cheng-Ming Chuong. 2013. “Feather Regeneration as a Model for Organogenesis.” Development, Growth and Differentiation 55, no. 1 (January): 139–148.
Lowe, Craig B., Julia A. Clarke, Allan J. Baker, David Haussler, and Scott V. Edwards. 2015. “Feather Development Genes and Associated Regulatory Innovation Predate the Origin of Dinosauria.” Molecular Biology and Evolution 32, no. 1 (January): 23–28.
McLain, M., M. Ross, M. Petrone, N. Lay, and M. Speights. 2023. “Response to “The Debate over Classification of Archaeopteryx as a Bird.” Answers Research Journal, in press.
Tay, Joel. 2019. “Feathered Pterosaurs: Ruffling the Feathers of Dinosaur Evolution.” Journal of Creation 33, no. 2 (August): 93–98.
Xu, Xing, and Y. Guo. 2009. “The Origin and Early Evolution of Feathers: Insights from Recent Paleontological and Neontological Data.” Vertebrata PalAsiatica 47, no. 4 (December 15): 311–329.
Yu, Mingke, Ping Wu, Randall B. Widelitz, and Cheng-Ming Chuong. 2002. “The Morphogenesis of Feathers.” Nature 420, no. 6913 (30 October): 308–312.
Supplementary Material
All supplementary data and figures are available on Zenodo at https://zenodo.org/record/7809453#.ZDCyNfbMLrc.








