The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
Crocodiles, alligators, caimans, and gharials are interesting reptiles that have key differences distinguishing them from all other reptiles. These animals can be classified into three families within the order Crocodylia: Alligatoridae (alligators and caimans), Crocodylidae (crocodiles), and Gavialidae (gharials). They are characterized by a long snout with sharp teeth, a long tail, sideways-protruding limbs, a thick scaly ectoderm, and a semi-aquatic, carnivorous lifestyle. As such, they seemingly belong to an apobaramin, separate from all other animals. How many kinds belong to Crocodylia? Are all crocodilian species interrelated, or do they possibly make up multiple, similar-looking kinds, like snakes? No data exists that suggests hybridization between the three crocodylian families therefore morphological analysis is uncertain. However, in this study, the mitochondrial DNA of 19 crocodilians as well as four outliers were aligned, and the species were clustered based on sequence similarity. The current results show a possible split between Alligatoridae and all other crocodilians, suggesting two crocodilian holobaramins. However, other lines of evidence need to be considered. The 37 genes in the mitochondrial genome have virtually the same order in the 19 crocodilians in the study, which may indicate that they are one group. Previous studies suggest three crocodilian holobaramins based on morphological cognita. However, other studies also found a different split between two groups: Alligatoroidea + Crocodyloidea, and Gavialoidea. When fossil species from Eusuchia were added, all of these species were found to form a single holobaramin. Thus, it may be possible that post-Flood extinctions caused the single Crocodylia holobaramin to fragment into subgroups.
Keywords: mitochondrial DNA; molecular baraminology; holobaramin; Crocodylia; Alligatoridae; Crocodylidae; Gavialidae
Introduction
The reptilian order Crocodylia is comprised of alligators, caimans, crocodiles, and gharials. These animals live in North and South America, Africa, India, Southeast Asia, and northern Australia. Crocodylia includes the families Alligatoridae, Crocodylidae, and Gavialidae. The latter two families belong to the clade Longirostres, since they have a longer, V-shaped jaw than alligators, which have a shorter, broader U-shaped jaw. Some researchers have placed the Malaysian false gharial (Tomistoma schlegelii) within Crocodylidae, whereas others have placed it within Gavialidae. All members of Crocodylia have hearts that are divided into two atria and two ventricles, as opposed to the hearts of all other reptiles, where there is a foramen between the two ventricles allowing oxygen-rich and oxygen-poor blood to mix (Nilsson 1994). Furthermore, these animals have a characteristic diapsid skull morphology with a long jaw and eyes placed at the top of the animal’s head.
Morphologically, crocodilians are distinct from all other reptiles, making them an apobaramin. For example, Frederico and McLain (2019) found discontinuity between crocodylians and non-crocodylian eusuchians based on their multidimensional (MDS) scaling plot and baraminic distance correlation (BDC), although it was weak. But do crocodilians form one or more holobaramins? The similar features of crocodiles, alligators, and gharials suggest that they might be able to form hybrids. As of yet, no such hybrids have been found, making it hard to discern if these species are all part of the same holobaramin.
But just because no hybrids have been discovered yet between the three crocodilian families does not mean that it is impossible, nor that hybridization did not happen in the past. Hennigan (2014) claims that it is probable that Crocodylia is a holobaramin, although he delineates crocodiles, alligators, and gharials as three separate kinds based on their strong cognita. These cognita include several morphological characteristics that allow researchers to distinguish between the four crocodilian groups, such as the shape of the snout, the location of sensory organs, the presence of salt glands on the tongue, and growth cycles (Britton 2013). God may have created multiple similar-looking crocodile holobaramins during Creation Week.
Therefore, in this study, the mitochondrial DNA (mtDNA) sequences of several species of Crocodylia were used in an attempt to clarify baraminic relationships exist in this order.
Materials and Methods
For the study of 19 crocodilian species, the mitochondrial DNA (mtDNA) was downloaded from the NCBI Organelle Genome website (n.d.). While the mtDNA represents only a very small fragment of the genome, the lack of recombination, high mutation rate, and absence of introns make it useful to work with (Gissi, Iannelli, and Pesole 2008).
Three species from the lizard family Lacertidae and also Sphenodon punctatus were used as outgroups. The heatmap and plots were created using R version 3.14. The ward.D2 clustering method was used to discover putative clusters. ClustalW (Thompson, Higgins, and Gibson 1994) was used to align the 19 crocodilian mtDNA sequences to form a sequence identity matrix. The Gene Content method (O’Micks 2017) was not performed for these species, since only three of them had complete proteomes at the UniProt website (n.d.).
The results of the analysis performed in this study can be found in Supplementary Files #1 online at Zenodo (2023) together with supplementary figures.
Results
The mtDNA similarity matrix has a Hopkins value of 0.848, which is good, indicating that there are real clusters (ostensibly baramins) in the data. The elbow plot in supplementary fig. 1 seems to indicate an optimal number of four clusters, which are all statistically significantly different at below the 1% level. At this value, the percent decrease in the ‘total within sum of squares’ (twss) values drops below 5%. The silhouette plot in supplementary fig. 2 also shows four optimal clusters. The heatmap showing the baraminic relationships between the 19 crocodilian species can be seen in fig. 1.
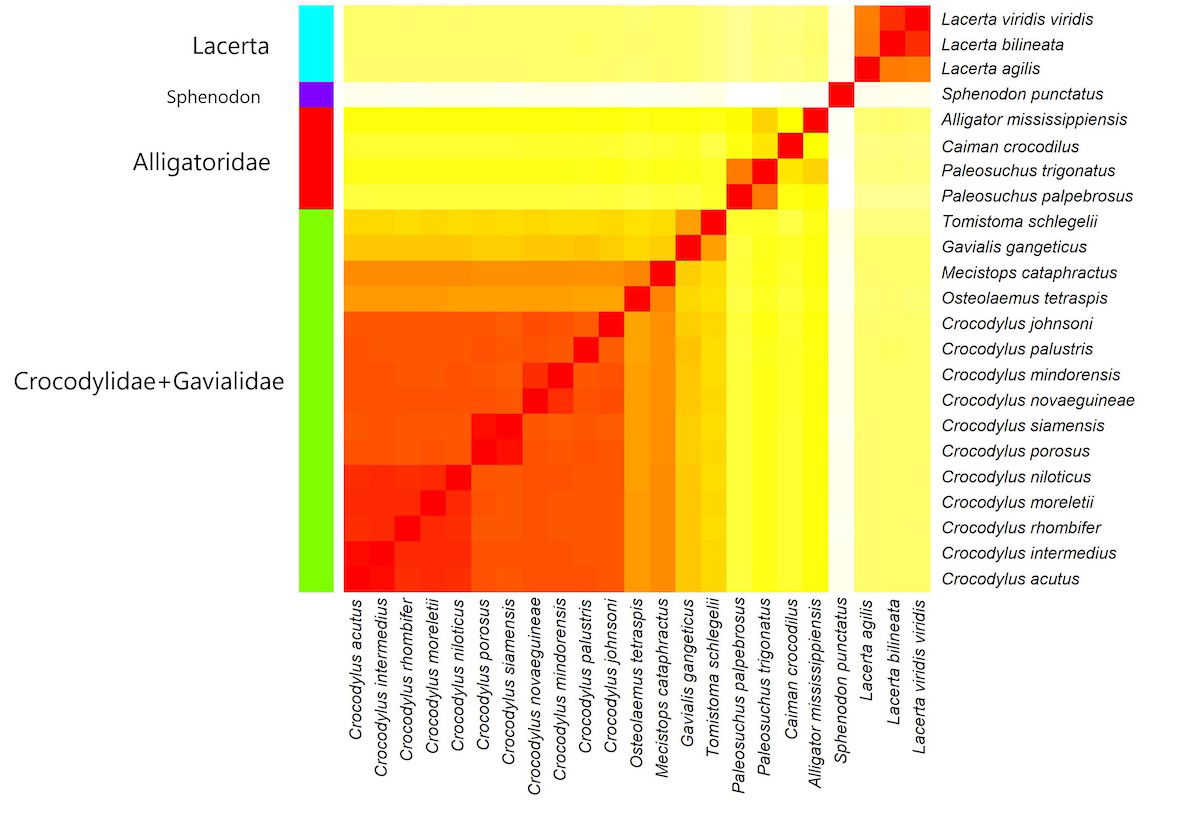
Fig. 1. Heatmap showing baraminic relationships between 19 crocodilian species, three Lacerta species, and S. punctatus based on mtDNA sequence similarity. Darker, redder colors denote high sequence similarity, indicating continuity between two species from the same holobaramin. Lighter, yellower colors denote low sequence similarity, indicating suggesting discontinuity between two species from different holobaramins.
The heatmap suggests that there are two crocodilian holobaramins: Alligatoridae (including the species Paleosuchus palpebrosus, Paleosuchus trigonatus, Caiman crocodilus, and Alligator mississippiensis) and Crocodylidae + Gavialidae (the 16 other crocodilian species). Lacerta and S. punctatus form a distinct outlier group and outlier species, respectively.
Four putative clusters can also be seen on the MDS plot in supplementary fig. 3. Whereas S. punctatus and the three Lacerta species are located in the upper left and the lower left corners, the situation with Crocodylidae + Gavialidae and Alligatoridae is more ambiguous. The four Alligatoridae species appear to be separate from the 15 Crocodylidae + Gavialidae species. The two Gavialidae species, Gavialis gangeticus and Tomistoma schlegelii are detached from the other 13 species within the group.
Species from Alligatoridae have shorter mtDNA (16,375 bp on average) with a higher average GC% (45.14% on average), whereas Crocodylia + Gavialidae have longer mtDNA (16,856 bp on average) and lower average GC% values (42.63% on average) (see table 1).
| Mean mtDNA length ± SD | Mean GC% ± SD | |
|---|---|---|
| Alligatoridae | 16375 ± 1245 bp | 45.14 ± 2.7% |
| Crocodilidae+Gavialidae | 16856 ± 175 bp | 42.63 ± 0.9% |
| Crocodylia | 16729 ± 629 bp | 43.16 ± 1.7% |
When the order of the 37 mitochondrial genes is examined, we can see that the gene order is uniform for all 19 species of Crocodylia (see fig. 2), and deviates only slightly in four genes (NAD4, trnH, trnS, and trnL) from the basic vertebral gene order (Montaña-Lozano et al. 2022). The only exception is Paleosuchus palpebrosus, which is missing the tRNA molecules for Thr, Pro, and Phe. The length of the mtDNA for this species is also shorter than the other 18, with a length of 14,935 bp, compared to the mean mtDNA length of 16,852 bp for all the other species.
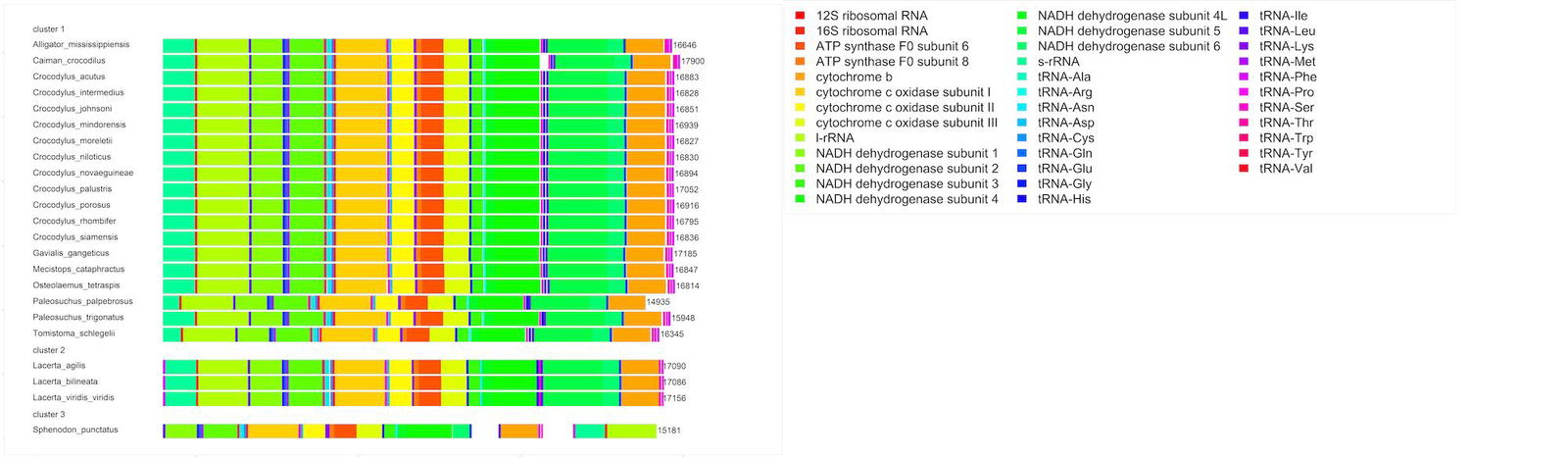
Fig. 2. Mitochondrial DNA gene map of the 23 reptile species from the Crocodylia study.
Discussion
How should we interpret these results? How does it compare to other studies involving Crocodylia? When we look at the mtDNA gene configuration we see that the gene order in all 19 Crocodylia species is almost the same, which may indicate one single holobaramin, but this is not conclusive. Furthermore, there are some differences between the mitochondrial gene order of crocodilians and other gnathostomes. For example, the tRNA molecules for Phe, Pro, and Thr are found 5’ of the mtDNA control region. Furthermore, the tRNA for His, Leu, and Ser also form a cluster, as opposed to all other vertebrates (Janke et al. 2005).
One could expect that if Alligatoridae and Crocodylidae + Gavialidae belong to separate holobaramins, they might have different mtDNA gene orders. However, the mtDNA gene order is also virtually the same within primates, which we know is made up of more than one holobaramin. The presence of several conserved spacer sequences in the mtDNA among all members of Crocodylia supports their being classified as a single holobaramin (Meganathan et al. 2011).
Interestingly, Frederico and McLain (2019) studied 80 morphological characters from 85 extant and extinct crocodilian species in a data set from Brochu (2011) using the BDISTMDS method (Wood 2008). They discovered four blocks of species, corresponding to Alligatoroidea, Crocodyloidea, Gavialoidea, and the non-crocodilian fossil Eusuchia. When only extant species were analyzed, their results seemed to indicate discontinuity between Alligatoroidea + Crocodyloidea, and Gavialoidea. Hennigan (2014) also classified Crocodylia into three groups, although he only studied extant species. However, when Frederico and McLain added fossil species from non-basal Euschia, this group connected Alligatoroidea + Crocodyloidea, and Gavialoidea, meaning that these groups likely form a contiguous holobaramin. Eusuchians are a group of extinct reptiles that resemble modern crocodilians, and are characterized by a secondary bony palate, allowing the animal to breathe through its nostrils while its head is submerged.
Besides the mtDNA we can also examine nuclear DNA. Oliveira et al. (2021) report that caimans (Caiman and Paleosuchus) have 21 pairs of nuclear chromosomes, yet alligators have 16 pairs. While it would be ideal if all species within the same holobaramin have the same number of chromosomes, this is not always the case, as numerous counterexamples abound. For example, within Macropodidae (kangaroos and wallabies), there is a wide range of karyotypes. The tammar wallaby (Macropus eugenii) has eight pairs of chromosomes, whereas the rock wallaby (genus Petrogale) has eleven (O’Neill et al. 1999). The mere fact that caimans and alligators have a different number of chromosome pairs does not exclude placing them in the same holobaramin. In general, the crocodilian karyotypes show low variability. As opposed to squamate reptiles, the tuatara, and turtles, crocodilian karyotypes have a small number of large chromosomes and numerous small-sized ones (Sriklunath, Thapana, and Muangmai 2015), which may indicate that Crocodylia is a single holobaramin.
Conclusion
The results coming from the mtDNA results alone suggest that there are four crocodilian groups. However, when the mtDNA results are combined with the morphological results from Frederico and McLain (2019), Crocodylia may be classified as one holobaramin. The addition of the fossil species could unite the groups into one. This is because different methodologies have failed to show consistent, holistic, discontinuity below the level of the order.
It may be that post-Flood extinctions caused the holobaramin Crocodylia to fragment into the several subgroups that we see today and that were detected by the mtDNA analysis. MtDNA has a tendency to split holobaramins into separate lineages. Since the mtDNA covers only a small fraction of the entire genome, further analysis (that is, of nuclear DNA) is warranted.
This study also highlights the importance of examining fossil taxa that may help fill in knowledge gaps when classifying species into holobaramins.
Lastly, since Crocodylia appears to form a holobaramin, this means that the kind can reach the level of the order. This has been proposed in other taxa, such as landfowl (Ahlquist and Lightner 2019).Though it may be a rule of thumb among baraminologists that the level of the kind is the family, this may vary, as we see here in the case of Crocodylia.
References
Ahlquist, Jon., and Jean K. Lightner. 2019. “Strategies for More Clearly Delineating, Characterizing, and Inferring the Natural History of Baramins I: Establishing Baraminic Status, with Application to the Order Galliformes (Class: Aves).” Creation Research Society Quarterly 56, no. 2 (January): 97–104.
Britton, A. 2013. “Crocodilians: Natural History and Conservation.” crocodilian.com.
Brochu, Christopher A. 2011. “Phylogenetic Relationships of Necrosuchus ionensis Simpson, 1937 and the Early History of Caimanines.” Zoological Journal of the Linnean Society 163, supplement 1 (December): 228–256.
Frederico, C., and M. A. McLain. 2019. “Baraminological Analysis of Fossil and Extant Eusuchia.” Journal of Creation Theology and Science Series B: Life Sciences 9: 1–2.
Gissi, C., F. Iannelli, and G. Pesole. 2008. “Evolution of the Mitochondrial Genome of Metazoa as Exemplified by Comparison of Congeneric Species.” Heredity 101: 301–320.
Hennigan, T. 2014. “An Initial Estimate toward Identifying and Numbering the Ark Turtle and Crocodile Kinds.” Answers Research Journal (2014):1–10. https://answersresearchjournal.org/ark-turtle-crocodile-kinds/
Janke, Axel, Anette Gullberg, Sandrine Hughes, Ramesh K. Aggarwal, and Ulfur Arnason. 2005. “Mitogenomic Analyses Place the Gharial (Gavialis gangeticus) on the Crocodile Tree and Provide pre-K/T Divergence Times for Most Crocodilians.” Journal of Molecular Evolution 61, no. 5 (November): 620–626.
Meganathan, P. R., Bhawna Dubey, Mark A. Batzer, David A. Ray, and Ikramul Haque. 2011. “Complete Mitochondrial Genome Sequences of Three Crocodylus Species and Their Comparison within the Order Crocodylia.” Gene 478, nos. 1–2 (June): 35–41.
Montaña-Lozano, Paula, Manuela Moreno-Carmona, Mauricio Ochoa-Capera, Natalia S. Medina, Jeffrey L. Boore, and Carlos F. Prada. 2022. “Comparative Genomic Analysis of Vertebrate Mitochondrial Reveals a Differential of Rearrangements Rate Between Taxonomic Class.” Scientific Reports 12, no. 1 (31 March): 5479.
NCBI Organelle Genome Database. n.d. https://www.ncbi.nlm.nih.gov/genome/browse#!/organelles.
Nilsson, S. 1994. “The Crocodilian Heart and Central Hemodynamics.” Cardioscience 5, no. 3 (September): 163–166.
Oliveira, Vanessa C. S., Marie Altmanová, Patrik F. Viana, Tariq Ezaz, Luiz A. C. Bertollo, Petra Ráb, Thomas Liehr, Ahmed Al-Rikabi, Eliana Feldberg, Terumi Hatanaka, Sebastian Scholz, Alexander Meurer, and Marcelo de Bello Cioffi. “Revisiting the Karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a Half-Century Delay: Bridging the Gap in the Chromosomal Evolution of Reptiles.” Cells 10, no. 6 (5 June): 1397.
O’Micks, Jean. 2017. “Baraminology Classification Based on Gene Content Similarity Measurement.” Creation Research Society Quarterly 54, no. 1 (Summer): 27–37.
O’Neill, R. J., M. D. Eldridge, R. Toder, M. A. Ferguson-Smith, P. C. O’Brien, and J. A. Graves. 1999. “Chromosome Evolution in Kangaroos (Marsupialia: Macropodidae): Cross Species Chromosome Painting Between the Tammar Wallaby and Rock Wallaby spp. with the 2n = 22 Ancestral Macropodid Karyotype.” Genome 42, no. 3 (June): 525–530.
Srikulnath, Kornsorn, Watcharaporn Thapana, and Narongrit Muangmai. 2015. “Role of Chromosome Changes in Crocodylus Evolution and Diversity.” Genomics and Informatics 13, no. 4 (December): 102–111.
Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. “CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment Through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice.” Nucleic Acids Research 122, no. 22 (November): 4673–4680.
UniProt Database. n.d. https://www.uniprot.org/proteomes.
Wood, Todd Charles. 2008. “Baraminic Distance, Bootstraps, and BDISTMDS.” Occasional Papers of the Baraminology Study Group 12: 1–17.
Zenodo. 2023. https://zenodo.org/record/7545965#.ZGThIqXMLrc.