The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
Lungfishes are a curiosity of nature. Like several modern fish species, lungfishes have lung-like structures in addition to their gills. Macroevolutionists claim that lungfishes represent a transition between sea and land organisms. However, despite their interesting organ, genomically, lungfishes cannot be described as a transitional species. The expression pattern of several genes in their lungs, vomeronasal organs (VNO), and limbs all differ from those in other fish analyzed. Among lungfish genomes, the 43 Gb genome of the Australian lungfish in particular is much larger than that of other known fish. Mitochondrial DNA analysis likely places lungfishes in their own separate apobaramin. Many modern fish are obligate air breathers, yet they are not on the alleged trajectory between sea and land organisms. Therefore, lungfishes are most likely just other species of fish with lungs and not transitional species that likely belong to their own separate kind(s).
Keywords: lungfish; tetrapods; baraminology; genome size; mitochondrial DNA
What kind of animals are lungfishes? Evolutionists point to these fascinating animals as the poster children of water-to-land evolution (Meyer et al. 2021; Schartl and Meyer 2021; Schartl et al. 2024). How should biblical creationists classify these animals? The fact that these unique animals have possible fish-like and amphibian characteristics makes them hard to classify. Perhaps, like the tuatara (Sphenodon punctatus) (Cserhati 2021) or the platypus (Ornithorhynchus anatinus), these animals may belong to their own separate kind(s).

Fig. 1. The Australian lungfish (Neoceratodus forsteri).
Extant lungfishes belong to the order Ceratodontiformes and live in various freshwater habitats, such as swamps, backwaters, and rivers. Four of the six extant species live in Africa (genus Protopterus), one lives in South America (Lepidosiren paradoxa), and one lives in Australia (Neoceratodus forsteri, see fig. 1). As their name suggests, lungfishes have both gills and lung-like structures (Kardong 2015, 421). These resemble the lungs of terrestrial animals with numerous septa and inner divisions including a ciliated epithelium (Icardo 2018). In contrast, bony fishes (class Osteichthyes) generally have sac-like gas bladders that only control buoyancy, whereas the lungs of lungfishes are subdivided into numerous air sacs. The African and South American lungfish species can also survive on dry land for up to a year by secreting a mucus-like substance around themselves that dries into cocoon-like membranes.
Baraminology is the study of taxonomy based on a biblical, historical interpretation of the book of Genesis. Genesis 1:20–21 says: “Then God said, ‘Let the waters abound with an abundance of living creatures, and let birds fly above the earth across the face of the firmament of the heavens.’ So God created great sea creatures and every living thing that moves, with which the waters abounded, according to their kind, and every winged bird according to its kind. And God saw that it was good.” A kind is a group of species that are continuous with one another and discontinuous with all other species. A kind is also called a ‘baramin,’ according to a widely used technical term. Baraminology is the study of these created kinds, and its goal is to determine the membership of these kinds.
The morphological and genetic characteristics of lungfishes will be compared to the characteristics of the amphibian genus Ambystoma (mole salamanders) in addition to a variety of selected fishes: Latimeria menadoensis (the Indonesian coelacanth), the lamprey family Petromyzontidae, and other fish to determine whether they belong to an existing kind or whether they form their own kind. More precisely, genetic characteristics of the mitochondrial genome (mtDNA) of lungfishes, as well as those from some of the previously listed species, will be compared to further clarify what kind lungfishes belong to or if they form their own kind.
Biblical Analysis
The Bible doesn’t specifically mention lungfishes. Due to their predominantly aquatic lifestyle, lungfishes were most likely created on Day Five of Creation Week with the other aquatic creatures (Genesis 1:20–21). Thus, they are separated from terrestrial animals created on Day Six. Lungfishes may survive on dry land, with some species able to for extended time periods. However, because lungfishes live primarily in water, they were likely created on Day Five and would not have been on the Ark during the Flood (Genesis 6:19–20).
Materials and Methods
A total of 77 mtDNA sequences were analyzed from the National Center for Biotechnology Information (NCBI) database from the six species of lungfishes as well as select mole salamanders, lampreys, eels, chimeras, sturgeons, gars, and coelacanths. These species are listed in Supplementary file 1 in the tab ‘Species.’ The West Indian Ocean coelacanth (Latimeria chalumnae) did not have an mtDNA sequence. The sequences were aligned in an all-versus-all pairwise manner using the ‘blseq’ Basic Local Alignment Search Tool (BLAST). Sequence similarity values from this alignment were assembled into a sequence identity matrix, which was visualized in a heatmap (fig. 2) using the ’heatmap’ method in R, version 4.3.1. Clustering was performed using the ‘average’ clustering method.
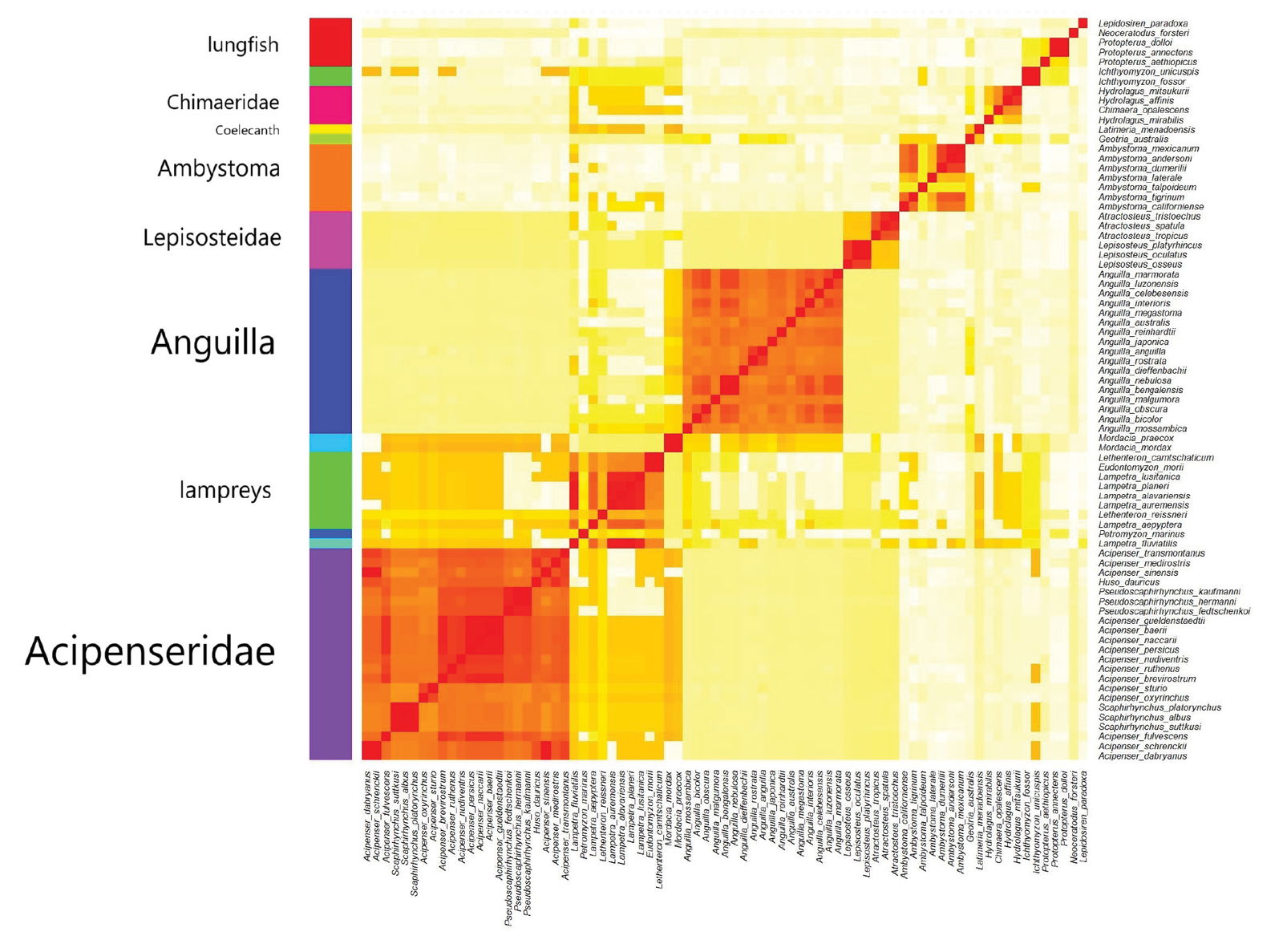
Fig. 2. Heatmap showing several putative holobaramins found in the mtDNA sequence comparison. Darker, redder colors denote species of the same kind, with higher sequence similarity values. Lighter, yellow colors denote species in separate kinds, with lower sequence similarity values.
Genome sizes and chromosome numbers were downloaded from the Genome Size Database (GSDB, Gregory 2024) and queried using the Genome Size Comparison + Analysis tool at the Molecular Baraminology Analysis Tool Suite (MBAT, https://molbar.shinyapps.io/molbar). They are available in Supplementary file 2 in the tab ‘Genome Size DB data.’ The ‘pairwise.t.test’ function in R was run on the C-values from the GSDB for the 11 families in the study to determine statistically significant p-values between pairs of families. The Elbow plot and the Silhouette plot are available in the Supplementary figures file. Supplementary files and figures are available on Zenodo at https://zenodo.org/records/14241580.
Results and Discussion
When it was first discovered, taxonomists thought the Australian lungfish was an amphibian (Krefft 1870). However, according to later evolutionary theory, lungfishes are more closely related to tetrapods (superclass Tetrapoda) than to ray-finned fish (class Actinopterygii) (Schartl et al. 2024; Wang et al. 2021). Based on the evolutionary timescale, lungfish fossils are dated at over 400 million years old, during what is called the Devonian period (Meyer et al. 2021), according to evolutionary geology, but which correspond to some of the earlier sedimentary layers laid down by the Flood. These fossils very much resemble modern lungfishes, meaning that lungfishes are considered living fossils. Due to their interesting respiratory and olfactory traits and limb structures, macroevolutionists claim that lungfishes are intermediate species between fish and terrestrial animals (Meyer et al. 2021; Wang et al. 2021).
A total of 374 species, including lungfishes and 49 families of teleosts (bony fish belonging to the infraclass Teleostei), in multiple ecological niches, exhibit various modes of air-breathing (Icardo 2018). Lungfishes can breathe air using their lungs. In comparison, some modern electric fish, which allegedly evolved after the transition of organisms from sea to land, obtain their oxygen via gills and air breathing. For example, the electric eel (Electrophorus electricus) is an obligate air breather, with richly vascularized oral mucosa. Air is gulped in through the mouth regularly at the surface of the water and then expelled through the operculum (Johansen et al. 1968; Kardong 2015, 418). Compared to other fishes, lungfishes alone have true lungs and a double circulatory system (consisting of a circuit carrying blood from the heart to the lungs and back to the body, plus another circuit carrying blood from the heart through the body and back to the heart) resembling that of tetrapods, with a mixing of oxygenated and deoxygenated blood (Icardo 2018).
Interestingly, evolutionists seemingly cannot decide whether the coelacanths or lungfishes are the closest extant relative of tetrapods. Numerous contradictions exist between morphological, molecular, and paleontological studies (Irisarri and Meyer 2016). This led Takezaki et al. (2004) to call finding the ancestor of tetrapods an “irresolvable trichotomy” between coelacanths, lungfishes, and tetrapods.
Phylogenetic trees between these three groups vary due to the choice of taxa, genes, and morphological characters analyzed. The classical view (fig. 3A) is that ceolecanths are a sister group to tetrapods. Some evolutionists place lungfishes next to tetrapods as a sister group based on the neighbor-joining and maximum-likelihood analysis of mitochondrial genomes (fig. 3B, Zardoya et al. 1998). Using the maximum-parsimony method to analyze the entire genome, Zardoya et al. (1998) place lungfishes (represented by P. dolloi) and coelacanths together as sister groups (fig. 3C). Trees based on various nuclear trees are contradictory. The tree seen in fig. 3A (where coelacanths are sisters to tetrapods) is supported by hemoglobin trees. The tree in fig. 2B (where lungfishes, represented by Protopterus are sisters to tetrapods) is supported by the amino acid sequences of DM20/PLP (Tohyama et al. 2001), RAG1, RAG2, POMC (Venkatesh, Erdmann, and Brenner, 2001). Lastly, the tree seen in fig. 2C (coelacanths and lungfishes are sisters to one another) is supported by the 28S ribosomal gene (Zardoya and Meyer, 1996).
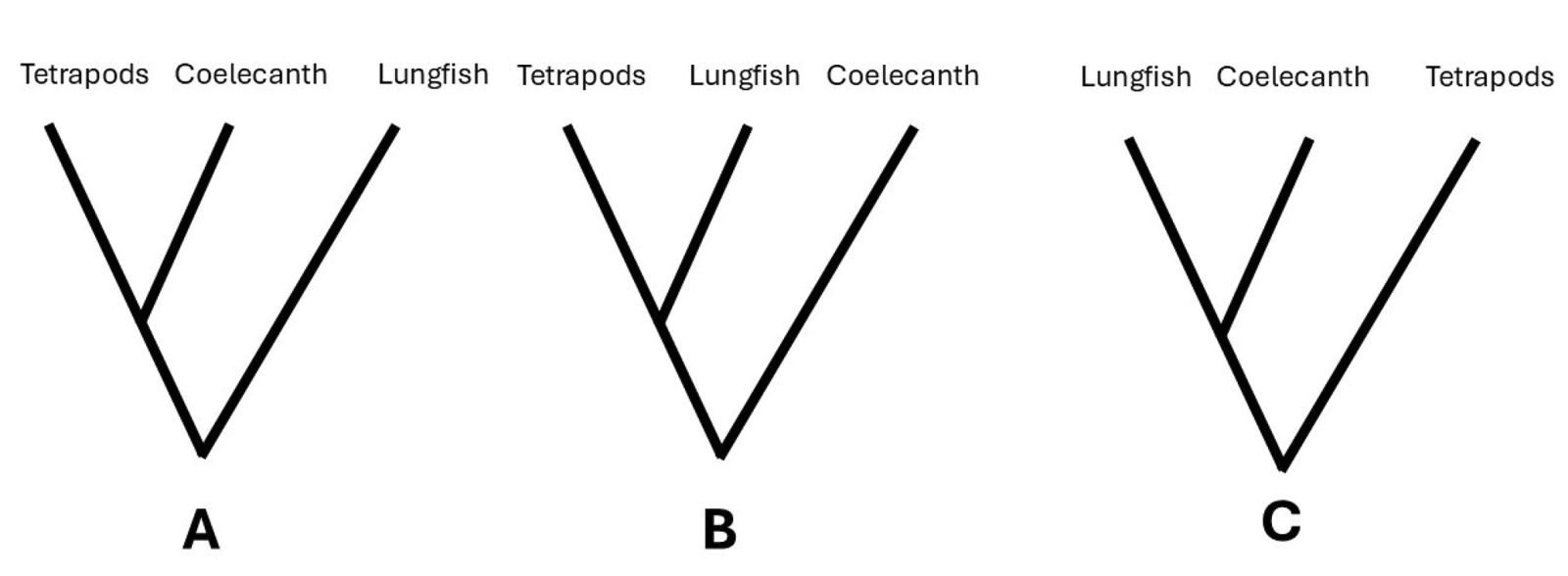
Fig. 3. Conflicting evolutionary trees of tetrapods, coelacanths, and lungfishes. A. The classical view that coelacanths are the sister group of tetrapods. B. The view that lungfishes are the sister group of tetrapods. C. Lungfishes and coelecanths as sister groups.
Alleged transitional characteristics
The presence, expression, and regulation of several genes in the Australian lungfish are different from those in all other fishes analyzed and are similar to the amphibians analyzed. For example, the lung of the Australian lungfish develops like amphibians (such as Xenopus laevis) where the shh (Sonic hedgehog) gene is heavily expressed in the embryonic lung (Yin et al. 2010). Furthermore, the number of genes in the surfactant B gene family is 2–3 times larger than that in all other fish. This gene lowers the surface tension of the lungs, thereby easing breathing and avoiding lung collapse (Meyer et al. 2021).
Olfactory receptor genes for detecting airborne odors are significantly more numerous in the Australian lungfish compared to other analyzed fishes, indicating that the Australian lungfish can survive for short periods of time on land. Conversely, the number of receptors for aquatic odors, such as eta and zeta receptors are smaller than in all other fish (Niimura and Nei 2005). The Australian lungfish also has a VNO like tetrapods, and the vomeronasal receptor gene family (such as the V2R genes) is greater than in all other fish (Meyer et al. 2021; Syed et al. 2017).
In the Australian lungfish, the Spalt Like Transcription Factor 1 (sall1) gene, which plays a role in the development of the distal part of the tetrapod limb (Sweetman and Münsterberg 2006) resembles that of tetrapods and has a similar function compared to the one in mice (Mus musculus). The presence and expression of four clusters of hox genes in the Australian lungfish (hoxa, hoxb, hoxc, and hoxd) are also similar to those of tetrapods. For example, the hoxc13 gene is also expressed in the limbs of the axolotl (Ambystoma mexicanum) (Meyer et al. 2021). For a summary of the role of these genes, see table 1.
Table 1. List of several differentially expressed genes between fish, tetrapods, and Neoceratodus forsteri (Meyer et al. 2021).
| Gene | Note |
|---|---|
| Eta, zeta olfactory receptors | Less in number than in fish |
| Surfactant protein B | Greater in number than in fish |
| Ssh | Expressed in embryonic lung |
| V2R genes | Greater in number than in fish |
| sall1 | Drives autopod development like in tetrapods |
| hoxc13 | Expression in distal limb similar to tetrapods |
The Australian lungfish genome
As a representative of lungfishes, the Australian lungfish genome is highly interesting. It has an estimated genome size of 43 gigabases (Gb), which is 30% larger than that of the axolotl (Nowoshilow et al. 2018). The genome sizes of other lungfishes are similar in size to each other (see table 2). An amazing 90% of the Australian lungfish genome is made up of repetitive DNA according to Meyer et al. (2021). Meyer’s team predicted 31,120 genes for the Australian lungfish genome, which matched 91.4% of the core vertebrate genes and 90.9% of the vertebrate conserved genes coming from the Benchmarking Universal Single-Copy Orthologs (BUSCO) database (Simão et al. 2015). Several characteristics of the Australian lungfish genome can be seen in table 3. In comparison, the African coelacanth genome is only 2.86 Gb in size and has 19,033 protein-coding genes. Its genome has a repeat content of only 25% (Amemiya et al., 2013). Based on the genome size alone, it is likely that the Australian lungfish is discontinuous with the coelacanth and the axolotl.
Table 2. List of genome sizes of several lungfish species.
| Latin name | English name | Genome size | Chromosomes |
|---|---|---|---|
| Neoceratodus forsteri | Australian lungfish | 43 Mb | 21 |
| Protopterus annectens | West African lungfish | 40.5 Mb | 27 |
| Lepidosiren paradoxa | South American lungfish | 46.2 Mb | 19 |
Table 3. Genome characteristics of the Neoceratodus forsteri (Meyer et al. 2021).
| Characteristic | N. forsteri |
|---|---|
| Estimated genome size | Approximately 43 Gb |
| Number of macrochromosomes | 17 |
| Number of microchromosomes | 10 |
| Number of genes | 31,120 |
| Average intron size | 50 Kb |
| Noncoding RNAs | 17,095 |
| Transfer RNAs | 1,042 |
| Ribosomal RNAs | 1,771 |
| Ribosomal RNAs | 3,974 |
| Repetitive DNA content | Approximately 90% |
Mitochondrial DNA analysis
The mtDNA for 77 species of lungfishes, amphibians, and various other fishes were downloaded and aligned. These included five species from the three extant families in the order Ceratodontiformes, seven species from the amphibian genus Ambystoma, 17 species of eels from the genus Anguilla, and 15 species of lampreys (class Hyperoartia); 14 from the Petromyzontidae family and one from the Geotriidae family. Four species from the family Chimaeridae (short-nosed chimaeras) were chosen to represent cartilaginous fishes (class Chondrichthyes), and 28 species from the class Actinopterygii were selected to represent ray-finned fishes. The latter consisted of 22 species from the family Acipenseridae (sturgeons) and six species from the family Lepisosteidae (gars). Lastly, the Indonesian coelacanth (Latimeria menadoensis) from the family Coelecanthidae was chosen as an outlier. Sarcopterygii, the clade or class (depending on the source) of lobe-finned fishes within the superclass Osteichthyes (bony fishes) to which both coelacanths and lungfishes belong, is likely higher than the boundary of the baramin. Thus, it is enough to use a species from another baramin as an outlier that is somewhat similar yet different.
The mtDNA sequences of these 77 species were aligned using the ‘blseq’ program, and the resulting sequence similarity matrix was visualized in a heatmap, seen in fig. 3. The Hopkins clustering statistic was 0.835, which corresponds to good clustering. The Elbow plot (Supplementary fig. 1) shows some distortion, so it is difficult to determine what the optimal cluster number is.
A total of 13 clusters were analyzed. Four groups had only one member. These are the Indonesian coelacanth (the outlier), the pouched lamprey (Geotria australis), the sea lamprey (Petromyzon marinus), and the European river lamprey (Lampetra fluviatilis). Because of their singleton species status, they were excluded from further analysis.
The sequence similarity clustering statistics in Supplementary file 1 (tab ‘statistics’) showed that the p-value was statistically significant (p < 5%) for five groups out of six that had at least three species. Only the lungfishes had a p-value (8%) above the cutoff limit of 5%. This could be because lungfishes belong to three families in the order Ceradontiformes. These are Ceratodontidae (the Australian lungfish), Lepidosirenidae (the South American lungfish), and Protopteridae (the four African lungfishes). While there are cases where species that belong to the same order have been assigned to the same holobaramin, such as the order Crocodilia (crocodilians) (Cserhati 2023), the kind boundary is often at the level of the family or the genus.
It seems that lungfishes form an apobaramin with multiple possible holobaramins. It could also be that lungfishes form a single holobaramin that has differentiated substantially over time. This could be because they now live in geographically disjunct areas (Australia, South America, and Africa). Since much time must necessarily have passed for the three lungfish families to reach their present geographical distribution, they may have undergone substantial diversification in the process. Lungfishes were likely not among the animal kinds present on Noah’s Ark, more than two of them may have survived the Flood, allowing for greater post-Flood diversification compared to Ark kinds.
Genome size and chromosome number comparison
Fig. 4 shows that the C-values for the three lungfish families are the largest across all eleven families studied. The C-value is the amount of physical DNA in picograms found in the haploid nucleus of a given organism. This value is the greatest for the South American lungfish, with a C-value range of 80.55–123.9 pg. This is about 60% larger than the C-values for the Australian lungfish (52.75–74.86 pg) or 18% larger than the C-values for the African lungfishes (genus Protopterus) (40.08–132.83 pg).
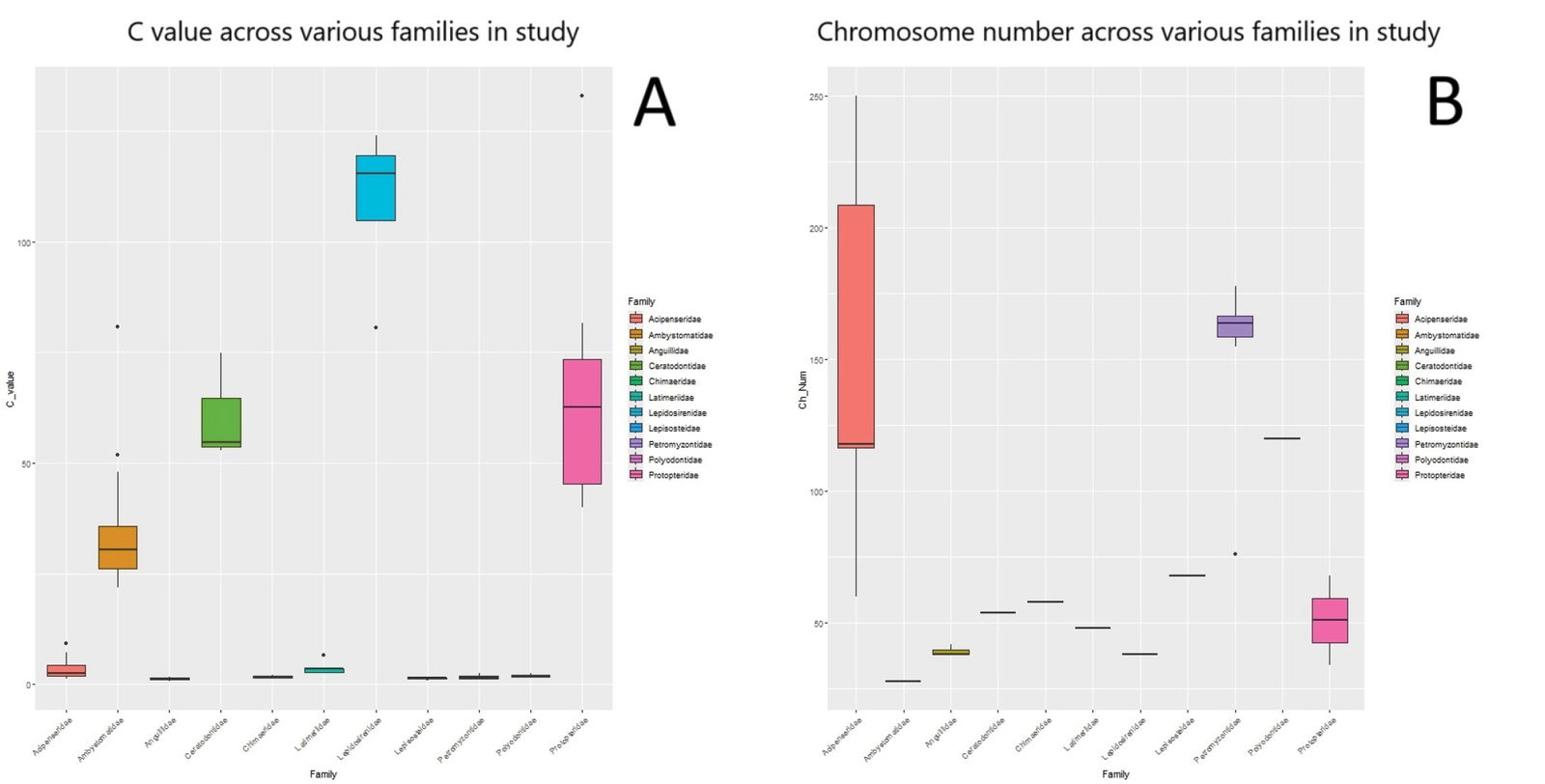
Fig. 4. The range of C-values across all 11 families used in this study. B. The range of chromosome numbers across all families in this study.
Unless the genome of the South American lungfish expanded due to the accumulation of repetitive sequences (which might be possible considering the high number of repetitive content of the Australian lungfish genome), it may be classified as its own holobaramin. The p-values between all three lungfish families in focus were statistically significantly different from all other families analyzed (Supplementary file 2, tab ‘pairwise t-test C-values’). The Australian lungfish (Ceratodontidae) and the African lungfishes (Protopteridae) have similar genome sizes, indicated by the p-value being 1.0.
South American lungfish have 38 chromosomes, while Australian lungfish have 54. African lungfishes have differing chromosome counts. West African lungfish (Protopterus annectens) have 34 chromosomes, while spotted lungfish (Protopterus dolloi) have 68, perhaps due to whole genome duplication. Though rare in animals (compared to plants), whole genome duplications are possible. For example, species in the bivalve family Mytilidae underwent whole genome duplications within the level of the kind (Corrochano-Fraile et al. 2022). Chromosome numbers may vary within kinds, such as in kangaroos and wallabies within the macropod kind (O’Neill et al. 1999), so it may be that South American lungfish still show continuity with the other two lungfish families.
Because of the very large size of the lungfish genomes compared to other species analyzed in this study, lungfishes are very likely a separate apobaramin. Their differing number of chromosomes may indicate the possibility of several lungfish holobaramins, but this is inconclusive. Similarly, even though snakes and lizards may look alike, it may be possible that a Common Designer created several snake and/or lizard kinds with common designs (Cserhati 2020). The case may be the same with lungfishes.
Summary and Conclusion
Macroevolutionists portray either lungfishes or coelacanths as transitional species between sea and land animals based on the presence of lungs, a double circulatory system, and the presence of VNOs. But the presence of these organs should not surprise biblical creationists. These organs, together with olfactory receptors that detect airborne odors, are complex organs. Evolutionists have not been able to present a series of mutations that transform air sacs into lungs, explain the formation of a double circulatory system from the single-looped system in other fishes, or describe the formation of the tetrapod-like VNO.
Rather, scientists should look at these organs as intelligently designed, irreducibly complex modular structures that God created in animals like fishes or amphibians. Most fish breathe with gills. Many also use air sacs for buoyancy. Some fish can also breathe using their lungs. The presence or absence of these organs in fish does not necessarily mean that one organ is derived from another. If fish did indeed evolve into land creatures, then this would have happened in the common ancestors of lungfishes and other fishes, which have not been found. Correlation doesn’t necessarily mean causation. The fact that lungfishes have both fish-like and tetrapod-like traits only reveals that life forms are very diverse.
Moreover, based on this analysis of the mitochondrial genome and size comparisons of the lungfishes’ nuclear genome with other fishes and amphibians, lungfishes probably do not belong to any other fish kinds, nor any amphibian kind. Lungfishes likely belong to their own separate kind(s), or possibly their own apobaramin. These interesting animals stand as a testimony to the creativity of our Creator.
References
Amemiya, Chris T., Jessica Alföldi, Alison P. Lee, Shaohua Fan, Hervé Philippe, Iain MacCallum, Ingo Braasch et al. 2013. “Analysis of the African Coelacanth Genome Sheds Light on Tetrapod Evolution.” Nature 496, no. 7445 (April 18): 311–316.
Corrochano-Fraile, Ana, Andrew Davie, Stefano Carboni, and Michaël Bekaert. 2022. “Evidence of Multiple Genome Duplication Events in Mytilus Evolution.” BMC Genomics 23, no. 1 (2 May): Article number 340. https://doi.org/10.1186/s12864-022-08575-9.
Cserhati, Matthew. 2020. “Do Lizards and Snakes Form Separate Apobaramins?” Journal of Creation 34, no. 1 (April): 96–101.
Cserhati, Matthew. 2021. “Baraminic Classification of the Tuatara (Sphenodon punctatus) Among Reptiles Based on Whole-Genome K-mer Signature Comparison.” Creation Research Society Quarterly 57, no. 3 (Winter): 201–205.
Cserhati, Matthew. 2023. “Baraminic Analysis of Crocodylia Based on Mitochondrial DNA Similarity.” Answers Research Journal 16 (May 31): 259–263.
Gregory, T. R. 2024. “Animal Genome Size Database.” http://www.genomesize.com
Icardo, José M. 2018. “Lungs and Gas Bladders: Morphological Insights.” Acta Histochemica 120, no. 7 (October): 605–612.
Irisarri, Iker, and Axel Meyer. 2016. “The Identification of the Closest Living Relative(s) of Tetrapods: Phylogenomic Lessons for Resolving Short Ancient Internodes.” Systematic Biology 65, no. 6 (November): 1057–1075.
Johansen, Kjell, Claude Lenfant, Knut Schmidt-Nielsen, and Jorge A. Petersen. 1968. “Gas Exchange and Control of Breathing in the Electric Eel, Electrophorus electricus.” Zeitschrift für vergleichende Physiologie 61 (June): 137–163.
Kardong, Kenneth V. 2015. Vertebrates: Comparative Anatomy, Function, Evolution. New York, New York: McGraw Hill.
Krefft, G. 1870. “Description of a Giant Amphibian Allied to the Genus Lepidosiren from the Wide Bay district, Queensland.” Proceedings of the Zoological Society of London 1870: 221–224.
Meyer, Axel, Siegfried Schloissnig, Paolo Franchini, Kang Du, Joost M. Woltering, Iker Irisarri, Wai Yee Wong, et al. 2021. “Giant Lungfish Genome Elucidates the Conquest of Land by Vertebrates.” Nature 590, no. 7845 (11 February), 284–289.
Niimura, Yoshihito, and Masatoshi Nei. 2005. “Evolutionary Dynamics of Olfactory Receptor Genes in Fishes and Tetrapods.” Proceedings of the National Academy of Sciences USA 102, no. 17 (April 11): 6039–6044.
Nowoshilow, Sergej, Siegfried Schloissnig, Ji-Feng Fei, Andreas Dahl, Andy W. C. Pang, Martin Pippel, Sylke Winkler, et al. 2018. “The Axolotl Genome and the Evolution of Key Tissue Formation Regulators.” Nature 554, no. 7690 (1 February): 50–55.
O’Neill, R. J., M. D. Eldridge, R. Toder, M. A. Ferguson-Smith, P. C. O’Brien, and J. A. Graves. 1999. “Chromosome Evolution in Kangaroos (Marsupialia: Macropodidae): Cross Species Chromosome Painting Between the Tammar Wallaby and Rock Wallaby spp. With the 2n = 22 Ancestral Macropodid Karyotype.” Genome 42, no. 3 (June), 525–530.
Schartl, Manfred, and Axel Meyer. 2021. “Neoceratodus forsteri (Australian lungfish).” Trends in Genetics 37, no. 6 (June): 600–601.
Schartl, Manfred, Joost M. Woltering, Iker Irisarri, Kang Du, Susanne Kneitz, Martin Pippel, Thomas Brown, et al. 2024. “The Genomes of All Lungfish Inform on Genome Expansion and Tetrapod Evolution.” Nature 634, no. 8032 (3 October): 96–103.
Simão, Felipe A., Robert M. Waterhouse, Panagiotis Ioannidis, Evgenia V. Kriventseva, and Evgeny M. Zdobnov. 2015. “BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs.” Bioinformatics 31, no. 19 (October 1): 3210–3212.
Sweetman, Dylan, and Andrea Münsterberg. 2006. “The Vertebrate spalt Genes in Development and Disease.” Developmental Biology 293, no. 2 (15 May): 285–293.
Syed, Adnan S., Alfredo Sansone, Thomas Hassenklöver, Ivan Manzini, and Sigrun I. Korsching. 2017. “Coordinated Shift of Olfactory Amino Acid Responses and V2R Expression to an Amphibian Water Nose During Metamorphosis.” Cellular and Molecular Life Sciences 74, no. 9 (May): 1711–1719.
Takezaki, Naoko, Felipe Figueroa, Zofia Zaleska-Rutczynska, Naoyuki Takahata, and Jan Klein. 2004. “The Phylogenetic Relationship of Tetrapod, Coelacanth, and Lungfish Revealed by the Sequences of Forty-Four Nuclear Genes.” Molecular Biology and Evolution 21, no. 8 (August): 1512–1524.
Tohyama, Y., H. Kasama-Yoshida, M. Sakuma, Y. Kobayashi, Y. Cao, M. Hasegawa, H. Kojima, Y. Tamai, M. Tanokura, and T. Kurihara. 1999. “Gene Structure and Amino Acid Sequence of Latimeria chalumnae (Coelacanth) Myelin DM20: Phylogenetic Relation of the Fish.” Neurochemical Research 24, no. 7 (July): 867–873.
Tohyama, Y., T. Ichimiya, H. Kasama-Yoshida, Y. Cao, M. Hasegawa, H. Kojima, Y. Tamai, and T. Kurihara. 2000. “Phylogenetic Relation of Lungfish Indicated By the Amino Acid Sequence of Myelin DM20.” Brain Research. Molecular Brain Research 80, no. 2 (September 15), 256–259.
Venkatesh, Byrappa, Mark V. Erdmann, and Sydney Brenner. 2001. “Molecular Synapomorphies Resolve Evolutionary Relationships of Extant Jawed Vertebrates.” Proceedings of the National Academy of Sciences USA 98, no. 20 (September 11), 11382–11387.
Wang, Kun, Jun Wang, Chenglong Zhu, Liandong Yang, Yandong Ren, Jue Ruan, Guangyi Fan, et al. 2021. “African Lungfish Genome Sheds Light on the Vertebrate Water-to-Land Transition.” Cell 184, no. 5 (March), 1362–1376.
Yin, Ao, Cecilia Lanny Winata, Svitlana Korzh, Vladimir Korzh, and Zhiyuan Gong. 2010. “Expression of Components of Wnt and Hedgehog Pathways in Different Tissue Layers During Lung Development in Xenopus laevis.” Gene Expression Patterns 10, nos. 7–8 (October–December): 338–344.
Zardoya, Rafael, Ying Cao, Masami Hasegawa, and Axel Meyer. 1998. “Searching For the Closest Living Relative(s) of Tetrapods Through Evolutionary Analyses of Mitochondrial and Nuclear data.” Molecular Biology and Evolution 15, no. 5 (May): 506–517.
Zardoya, Rafael, and Axel Meyer. 1996. “Evolutionary Relationships of the Coelacanth, Lungfishes, and Tetrapods Based on the 28S Ribosomal RNA Gene.” Proceedings of the National Academy of Sciences USA 93, no. 11 (May 28), 5449–5454.