The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
There are 31 xenarthran species living today, made up of armadillos, anteaters, and sloths. These animals are characterized by a narrow, elongated skull and a unique vertebral process in the posterior trunk. Xenarthra represents an apobaramin within which several holobaramins may be discerned, using both mitochondrial DNA and morphological data. On a biblical basis, armadillos could possibly be classified as remes (creeping) animals because of their burrowing activity. On the other hand, sloths and anteaters are likely beasts of the field (b’hemah) because of their larger size and movement and the arboreal lifestyle of some sloths. The morphological results suggest that there are two armadillo holobaramins. The first group is made up of genera, such as Dasypus, Tolypeutes, Kuntinaru, Stegotherium, Priodontes, and Cabassous. The other group consists of the genera Peltephilus, Vassallia, Propalaeohoplophorus, Eutatus, Chlamyphorus, Zaedyus, Prozaedyus, Paleuphractus, Macroeuphractus, Doellotatus, Proeutatus, Euphractus, and Chaetophractus. The mitochondrial DNA analysis shows that Cabassous and Tolypeutes separate from Dasypus. However, this may be due to the fragmentary nature of the mitochondrial genome. It appears that Vermilingua (anteaters) approximate a single holobaramin, consisting of the genera Tamandua, Myrmecophaga, and Cyclopes. This can be seen based on the mitochondrial DNA results. There is one instance of morphological continuity between Tamandua and Priodontes, but Tamandua is also discontinuous with most genera in the large second group of armadillos. Concerning sloths, it appears that there may be one or more groups. The two extant genera, Bradypus and Choloepus are tree sloths, which might be separate from ground sloths due to their locomotion, although there is no decisive evidence for discontinuity between tree and ground sloths. On a molecular level, however, all sloths share numerous “sloth endogenized foamy-like retroviruses,” which unite them. Furthermore, the dental formula of 5+4 upper and lower teeth is generally characteristic of all sloths.
Keywords: Xenarthra, Cingulata, Pilosa, Vermilingua, Dasypus, mitochondrial DNA, BARCLAY, molecular baraminology
Introduction
Xenarthrans are animals with elongated skulls, small brains, and a unique vertebral process called the xenarthrous articulation, present in the posterior portion of the trunk that aids in digging (Oliver et al. 2016). Furthermore, they have a low body temperature, low metabolic rate, and intra-abdominal testes. They are separate from all other mammals because of their reduced dentition, along with reduced enamel (Gaudin and Croft 2015). They are so different from other mammals, that it was suggested that they belong to their own group besides all other placentals (Delsuc et al. 2002; Novacek 1992). Based on these considerations, these animals likely form an apobaramin.
Superorder Xenarthra is made up of two orders, Cingulata (armadillos) and Pilosa (hairy xenarthrans), itself made up of two suborders, Folivora (sloths) and Vermilingua (anteaters). The NCBI database (https://www.ncbi.nlm.nih.gov/nucleotide) contains data for 37 xenarthran species, although most authors count 31 extant species: namely 21 armadillos, four anteaters, and six sloths, which live mainly in North and South America (Anderson and Handley 2001).
Armadillos are a group of omnivorous and insectivorous animals with flexible body armour. An extinct group of large-sized armadillos, called glyptodonts are, according to Mitchell et al. (2016) closely related to the subfamily Euphractinae (six-banded, dwarf, and hairy armadillos). Extant Cingulata can be divided into two families, Dasypodidae and Chlamyphoridae, the former made up of a single genus, Dasypus, whereas the latter is made up of three subfamilies, Chlamyphorinae (fairy armadillos), Euphractinae, and Tolypeutinae (naked-tailed, giant, and three-banded armadillos) (Gibb et al. 2015).
Anteaters are small to large-sized animals that are specialized in feeding on insects, such as ants and termites. They are characterized by long, thin toothless snouts, with a thin, flat saliva-coated tongue used to catch its insect prey. Hence the name Vermilingua, or “worm tongue.” The front feet have large claws on the third digit, which the animal uses to break down ant and termite mounds. The body has dense fur, and the tail is long.
There are various types of sloths, classified based on their lifestyle. Some authors distinguish between two genera of tree sloths: the three-toed sloths (Bradypus) and the two-toed sloths (Choloepus) (Gaudin 2004). Tree sloths are medium-sized folivores that live in trees and can hang upside down with their rigid hook-like claws. Ground sloths live on the ground and are either herbivores or omnivores. Only one species of sloth lives in the water, Thalassocnus, which is extinct.
Various authors differ as to which groups are related to one another, and which groups are monophyletic, such as Dasypus, Euphractinae (Euphractus, Chaetophractus, and Zaedyus), or Tolypeutinae (Tolypeutus, Cabassous, and Priodontes) (Billet et al. 2011; Möller-Krull et al. 2007). Resolving the baraminic relationships between Cingulata and other xenarthrans is a worthwhile pursuit. Xenarthra probably represents an apobaramin consisting of at least three groups, within which individual holobaramins and monobaramins may be discerned. Besides the 37 extant species, 200 extinct genera of Xenarthra have been noted (Möller-Krull et al. 2007).
Interestingly, mitochondrial DNA has been isolated from a species in the glyptodont genus Doedicurus (Mitchell et al. 2016). Ancient DNA from these species might help resolve baraminic relationships between extant and extinct armadillo species. Ancient DNA has also been isolated from two extinct ground sloth species, the Shasta sloth (Nothrotheriops shastensis), and Megatherium americanum, which both come from the Family Megatheriidae. Poinar et al. (2003) found that these two ground sloth species were most similar to two-toed sloths (Megalonychidae) based on the alignment of three concatenated nuclear genes, the von Willebrand Factor (vWF), the cAMP-responsive element modulator gene (CREM), and phospholipase C, β 4 (PCB4).
Ancient DNA has been analyzed in previous baraminology studies. For example, Cserhati (2023) amalyzed mitochondrial DNA (mtDNA) from Homo heidelbergensis to classify it as a member of the human holobaramin.
The purpose of this baraminology study is to analyze baraminic relationships within Xenarthra based on mtDNA sequence similarities, and two morphological character sets. Xenarthrans probably represent an apobaramin, and it will be worth determining what holobaramins and monobaramins are present in this group of animals.
Materials and Methods
Sequence data
The mtDNA sequences of 35 extant and 2 extinct Xenarthra species were downloaded from the Organelle Genome Browser at the NCBI Database website at https://www.ncbi.nlm.nih.gov/genome/organelle. One ancient DNA sequence was downloaded from the Nucleotide Database at NCBI, belonging to Doedicurus sp. (accession number KX098449).
This particular 15,936 bp long sequence consisted of 44.9% undetermined base pairs (Ns). In order to retain this sequence in the data set, those portions of the multiple alignment created from the mtDNA sequences were cut out for all species where there were Ns at those positions. Delsuc et al. (2001) also removed indels and hypervariable regions from three gene regions (ADRA2B, BRCA1, and vWF) that they analyzed to study phylogenetic relationships within Xenarthra.
The analysis with these self-same sequences was also carried out without making the cuts. This was done to verify that making the cuts does not change the clustering results.
Morphological character sets
A morphological data set containing 125 characters for 22 extant and extinct armadillo species was taken from Billet et al. (2011). Character states were coded from 0–4, and missing values were represented with a ‘?’ symbol. Distance Correlation Analysis (DCA) and Classic Multidimensional Scaling (MDS) were used to analyze this data set. Pearson correlation and baraminic distance were selected, and a minimum character relevance cutoff value of 0.75 was chosen, to retain as many characters as possible. After this cut, 99 characters remained (79.2% of the original 125). The Partition Around Medoids (PAM) algorithm was used to generate the Silhouette plot, seen in Supplementary fig. 1.
A second morphological data set containing 225 craniodental characters for 46 xenarthran species (armadillos and sloths) was taken from Appendix 3 of Gaudin (2004). Several taxa had to be excluded, since a large proportion (> 15%) of their characters were undefined or missing (‘?’), these include Cyclopes, Myrmecophaga, Palaeomyrmidon, Pholidota, Protamandua, and Tamandua. Glossotherium was also removed because this taxon did not have all 225 characters in the data set. Distance Correlation Analysis (DCA) and Classic Multidimensional Scaling (MDS) were used to analyze this data set as well. Pearson correlation was selected, and a minimum character relevance cutoff value of 0.75 was chosen, after which 169 characters remained (75.1% of the original 225). The Partition Around Medoids (PAM) algorithm was used to generate the Silhouette plot, seen in Supplementary figs. 1 and 2.
Software
The 38 mtDNA sequences were aligned using the ClustalW software. Heatmaps were created using the “single” clustering algorithm using the “heatmap” function in R. The elbow plot was created using the ‘fviz_nbclust’ function. The Gap Statistic method was employed to estimate the optimal number of clusters using the ‘clusGap’ command in R using the ‘kmeans’ clustering algorithm. R version 4.1.0. was used to analyze the data and generate plots.
The Billet et al. (2011) and Gaudin (2004) data sets were analyzed using the Baraminology and Cluster Analysis (BARCLAY) software available at https://coresci.org/barclay (Wood 2020). The Kinemage, Next Generation (KING) software was used to generate the multidimensional scaling (3D MDS) plot (Chen, Davis, and Richardson 2009).
Supplementary data availability
The supplementary figures and files generated during this analysis can be found online at the Zenodo website at https://zenodo.org/record/11671349.
Results
Morphological analysis
The Billet et al. (2011) data set was analyzed using the BARCLAY algorithm, as described in the Materials and Methods section. The average silhouette width for PAM analysis with two clusters was 0.25, compared to 0.19 for three clusters, 0.17 for four, and only 0.09 for five clusters (Supplementary fig. 1). The Hopkins statistic was only 0.687, which denotes fair clustering. Thus, two putative baramins were assumed in the rest of the analysis. The baraminic distance plot can be seen in fig. 1, and the corresponding multidimensional scaling plot can be seen in fig. 2. Additional results from the BARCLAY analysis can be found in Supplementary File 1 online.
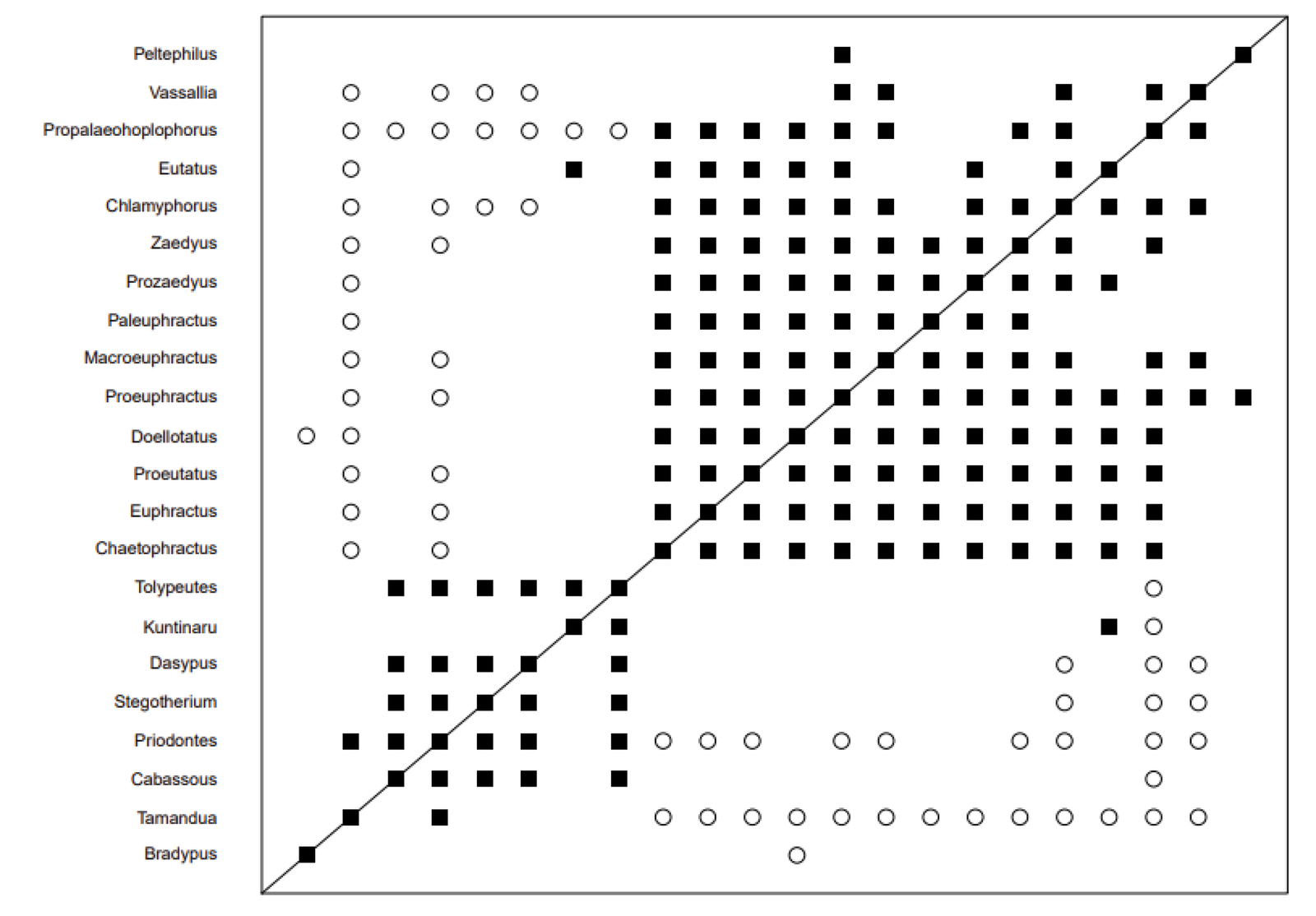
Fig. 1. Baraminic distance plot based on the armadillo character matrix from the Billet et al. (2011) data set. A larger cluster of 14 armadillo species can be seen in the upper right. A smaller group of six armadillo species separated by significant discontinuity can be seen in the lower left. There is one instance of morphological continuity between Tamandua (Vermilingua) and Priodontes, but Tamandua is also discontinuous with most genera in the large second group of armadillos. Bradypus (Folivora) lack continuity with all armadillos and shows discontinuity with one of them (Doellotatus).
What these figures show is that the 22 species can be divided into two putative holobaramins, a larger one of 14 species (Peltephilus, Vassallia, Propalaeohaplophorus, Eutatus, Chlamyphorus, Zaedyus, Prozaedyus, Paleuphractus, Macroeuphractus, Proeuphractus, Doelotattus, Proeutatus, Euphractus, and Chaetophractus), and a smaller one of only six (Tolypeutes, Kuntinaru, Dasypus, Stegotherium, Priodontes, Cabassous). These two groups largely show continuity within themselves and a substantial amount of discontinuity between them. Furthermore, Bradypus (sloth) does not appear to group with either the larger or the smaller cluster but shows discontinuity with Doellotatus from cluster 1. There is only one instance of continuity between Tamandua (anteater) and Priodontes from the smaller cluster, but Tamandua is also discontinuous with most genera in the larger cluster of armadillos. Thus, Bradypus and Tamandua probably belong to separate groups.
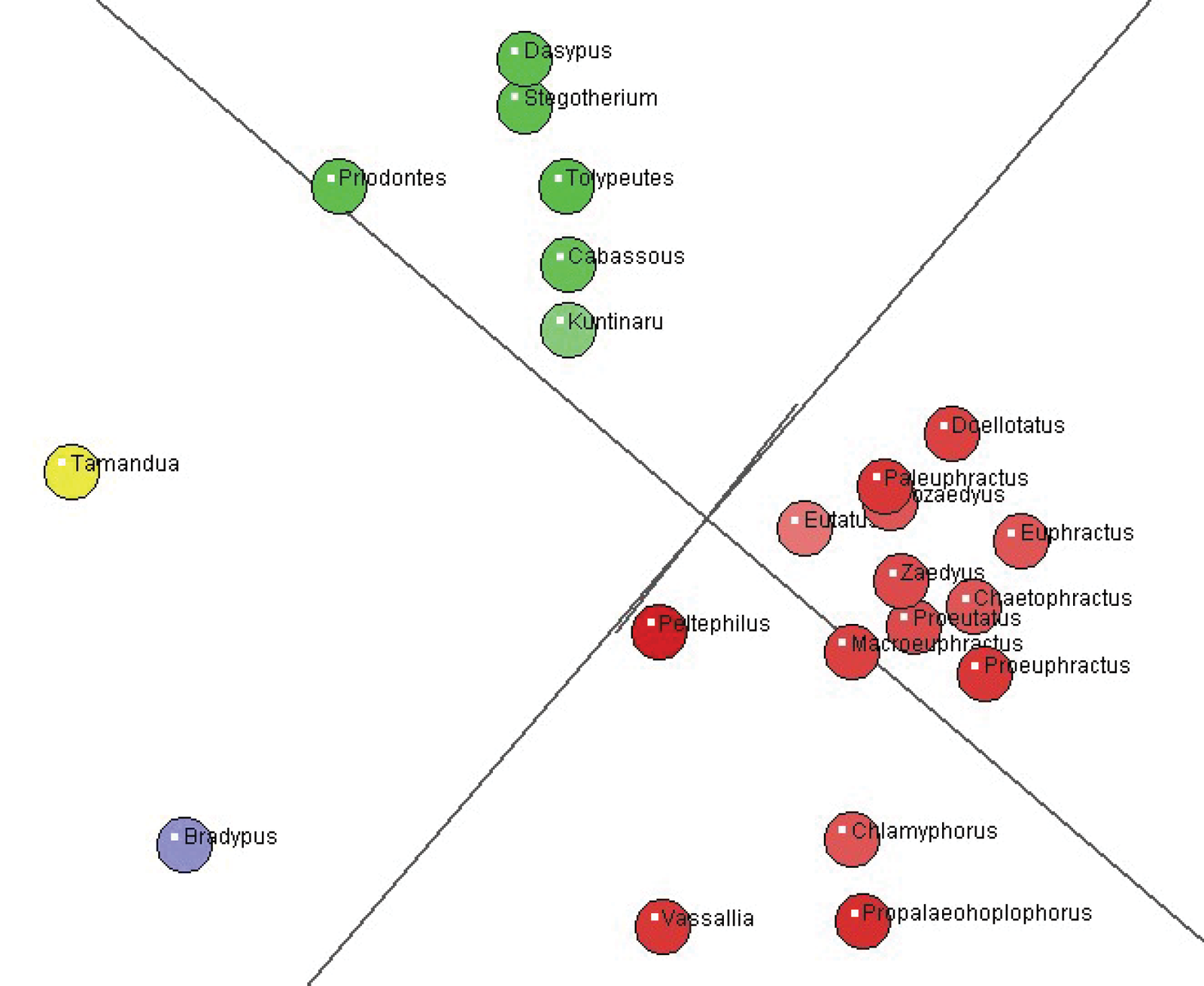
Fig. 2. Multi-Dimensional Scaling (MDS) plot of two armadillo groups (red and green), with 14 and six species, respectively. Tamandua (Vermilingua) is in yellow and Bradypus (Folivora) in blue.
In fig. 2 we see that the larger and smaller armadillo clusters separate from one another quite well in the multidimensional scaling analysis. The position of Eutatus may be somewhat ambiguous (somewhere in the morphospace between the two clusters). This may be because this taxon shows continuity with Kuntinaru in the baraminic distance analysis (fig. 1). Its significant correlation is 0.454 with Kuntinaru, whereas it shows no correlation with the rest of the smaller cluster.
The Gaudin data set was analyzed with the BARCLAY algorithm. The results can be found in Supplementary File 2 online. The Silhouette plot can be seen in Supplementary fig. 2, and the baraminic distance plot can be seen in fig. 3. It is somewhat ambiguous as to whether there are two or three clusters in the data. The average silhouette width was 0.33 for three clusters, whereas it was 0.35 for two. Therefore, the rest of the analysis was performed on two clusters.
One group is very visible in fig. 3, whereas the second larger group appears to be less well-formed. Extant species have their Latin names in all capital letters.

Fig. 3. Baraminic distance plot based on the armadillo and sloth character matrix from the Gaudin (2004) data set.
The smaller, better-formed group consisting of seven species in the upper right includes species from Cingulata. In the bottom left, there are 32 species, which make up the less well-formed larger group. These 32 species appear to form two possible monobaramins. Ten of the larger group of 32 species are extinct sloths, which seem to form a monobaramin. These belong to the families Mylodontidae and Scelidotheriidae.
The remaining 22 species include both ground sloths and tree sloths. Here, the two extant sloth groups, Choloepus (two-toed sloths) and Bradypus (three-toed sloths) belong to this same larger group, indicating that since there is continuity between them, then they must belong to the same holobaramin. The 32 sloth species are very spread out in the principal component analysis (PCA) plot in Supplementary fig. 2.
The reason it may seem that there are two separate monobaramins within the larger sloth holobaramin of 32 species is that seven of these species are extinct species. Only Nematherium from the smaller sloth group of ten species shows significant continuity with seven species from the larger sloth group in the lower left of fig. 3. Conversely, only Octomylodon from the larger sloth group shows significant continuity with five species from the smaller group of sloths.
Fig. 4 shows the multidimensional scaling plot showing the 39 species analyzed in the Gaudin data set. There is a clear separation between the armadillos and the sloths. However, as can be seen in Supplementary fig. 3, the two putative sloth monobaramins also seem to separate from one another.
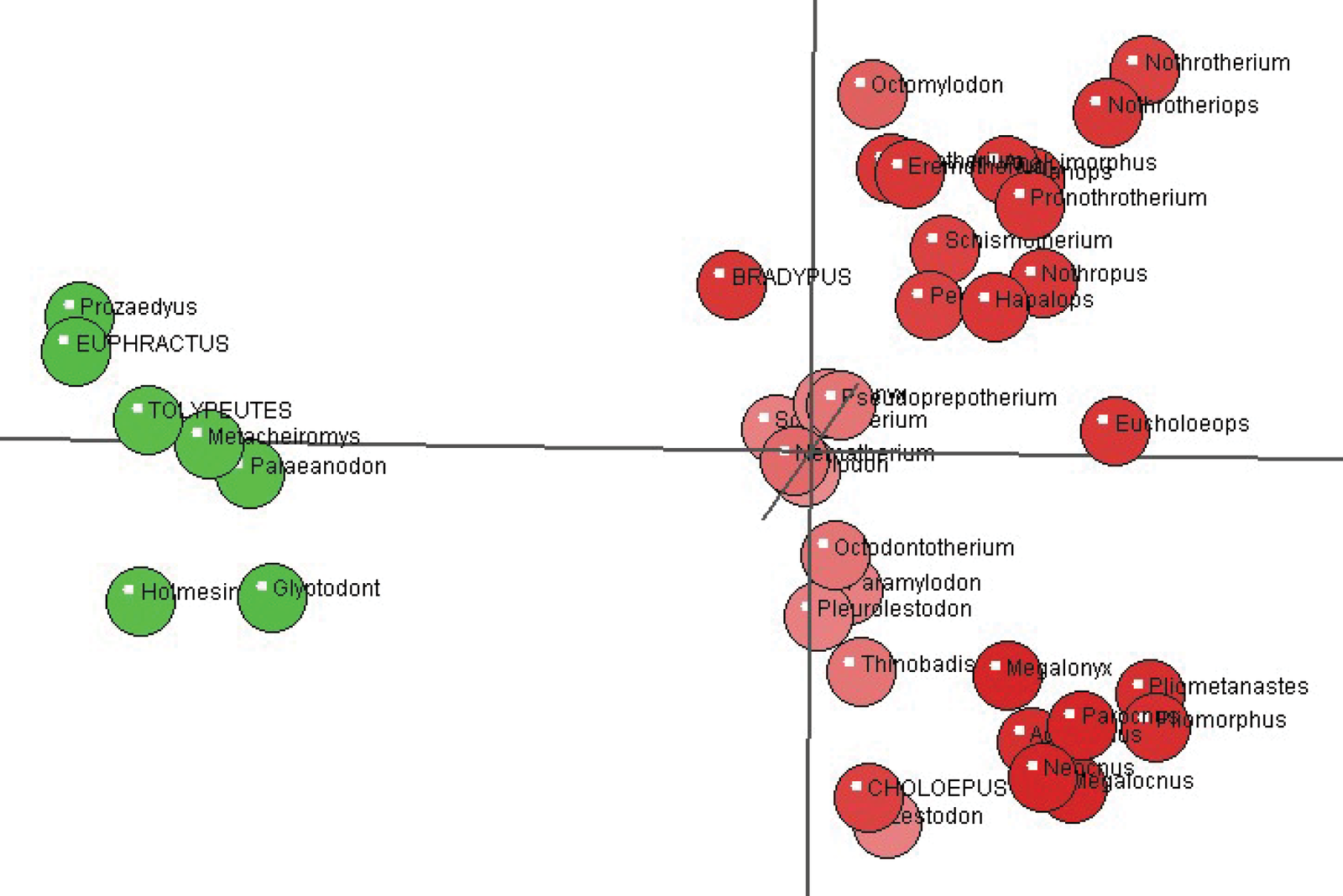
Fig. 4. Multi-Dimensional Scaling (MDS) plot of an armadillo and a sloth group. The sloth group consists of 32 species (red), whereas the armadillo group is made up of seven species (green).
Mitochondrial DNA analysis
For the mtDNA analysis, the mitogenomes of 38 species were aligned, including 37 xenarthrans and Monodelphis domestica (opossum). After removing the undefined bases from the alignment caused by Doedicurus, a sequence identity matrix was calculated. The results of this analysis can be found in Supplementary File 2. The Hopkins clustering value is 0.789, which indicates good clustering.
The Hopkins clustering value calculated on the alignment without cutting out the polyN stretches of DNA from the mtDNA sequence of Doedicurus was 0.844. The clustering results were the same irrespective of whether the polyN DNA segments were cut or not.
Based on mtDNA and nuclear evidence analyzed by Delsuc, Stanhope, and Douzery (2003) and Delsuc et al. (2012), three armadillo baramins may exist: namely Dasypodinae, Tolypeutinae, and Euphractinae. When analyzing the mtDNA data, there appeared to be four to six possible clusters in the data. Cluster membership and statistics were calculated for 4–6 clusters. The analysis began with a minimum of four clusters since there were three groups (armadillos, anteaters, sloths) and opossum as an outlier species. The elbow plot in Supplementary fig. 4 shows that the total within sum of squares (twss) value stops decreasing sharply at six clusters. However, the silhouette plot shown in Supplementary fig. 5 shows a maximum average silhouette width at five clusters.
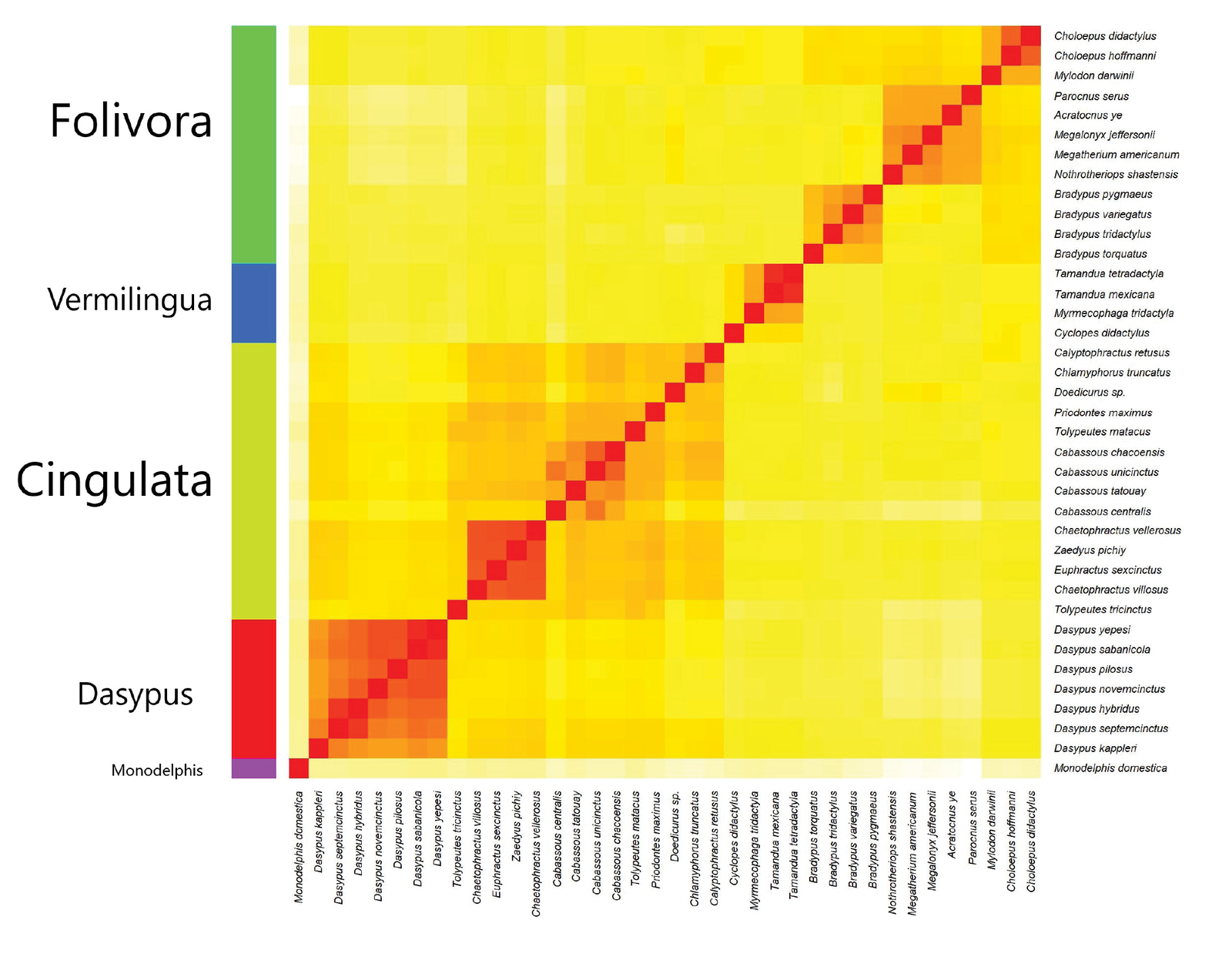
Fig. 5. Heatmap based on sequence similarity between 37 Xenarthra species and the outlier species, Monodelphis domestica (opossum). The 37 Xenarthra species form four groups: green: Folivora (sloths), blue: Vermilingua (anteaters), yellow: Cingulata (non-Dasypus armadillos), and red: Dasypus (armadillos). Darker, redder colors represent species pairs with higher mtDNA sequence similarity (and thus belong to the same kind). Yellow, brighter colors represent species from two separate kinds (which correspond to lower mtDNA sequence similarity).
When compared to the morphological data, we see that the placement of the sloth genus Bradypus is rather ambiguous. It does not show continuity with any other genus and shows discontinuity with Doellotatus (see fig. 1). Thus, the baraminic placement of Bradypus is unsure. It could either be a sloth monobaramin, or its own holobaramin. Collagen-based evidence also placed Bradypus together with Nothrotheriops (Presslee et al. 2019). This, however, is discordant with the mtDNA results.
Fig. 5 shows the heatmap of the 37 Xenarthra species with five clusters (the four xenarthran groups and opossum as the outlier species). Cingulata is subdivided into two groups: the genus Dasypus, with seven species, and all other cingulates (14 other species). In contrast with the morphological results, Tolypeutes and Cabassous separate from Dasypus.
Furthermore, Doedicurus is nested within all other non-Dasypus armadillos, indicating continuity between extant and extinct cingulates (Delsuc at el. 2016). This species is closest to Chlamyphorus truncatus (the pink fairy armadillo) with an mtDNA sequence similarity of 0.832 and Calyptophractus retusus (the greater fairy armadillo) with a similarity value of 0.828. It is possible that small-sized fairy armadillos represent a monobaramin within which species underwent size reduction, enlargement of digging claws for a subterranean lifestyle, accompanied by reduced eyes (Möller-Krull et al. 2007).
Discussion
Vermilingua
It appears that there is only one Vermilingua holobaramin, as shown by the mtDNA evidence. This means that all anteaters belong to the same created kind. However, it appears that there are two Cingulata holobaramins: Dasypus and euphractine armadillos. This is supported by both the morphological and the molecular evidence. As to why there are multiple armadillo baramins, we cannot be sure. It may have been the case that God created multiple armadillo kinds, just as there appear to be multiple bat and snake kinds (Cserhati 2020; Cserhati 2021).
Folivora
With Folivora, there might be evidence for the holobaraminic status of Bradypus, although this is contradicted by the craniodental evidence from Gaudin (2004), where Bradypus is placed into the same group as Choloepus. Interestingly, Gaudin (1995) also notes that the two tree sloths share some similarities with one another that set them apart from other sloths, most notably their suspensory locomotion.
The craniodental evidence may support the existence of two sloth monobaramins, although this is inconclusive. Due to the lack of discontinuity between the two groups, it cannot be said with confidence that they belong to two separate holobaramins. Thus, they appear to be two monobaramins within a larger sloth holobaramin. In fact, a molecular marker, a 27 bp deletion was detected in Bradypodidae (where Bradypus belongs) and Megalonychidae (where Choloepus belongs), but not in Vermilingua. Furthermore, eight retroposons (3× DAS-Ia, 2× DAS-IIa, DAS-III1b, U2, L1MB2) and 16 indels were present only in Dasypus (Möller-Krull et al. 2007), evidence for discontinuity between that group and other armadillos.
Bradypus and Choloepus are the only two extant sloth genera. Dissimilarities may have arisen in the mtDNA of Bradypus ever since they diverged from extinct sloths, which were highly diverse, including around 100 genera in the past. Thus, the two extant genera might have been the leftovers of massive extinction around the time of the Flood. Bradypus happens to have many autapomorphic morphological character states. However, these characters are also found in genera from Megatheriidae, namely Planops, Megatherium, and Eremotherium (Slater et al. 2016). Slater et al. also found several sloth endogenized foamy-like retroviruses (SloEFV) that are present in all sloth lineages, and that Choloepus contains certain SloEFVs not found in other sloths, indicating that it may be a monobaramin. Additionally, the dental formula of sloths is conservative, corresponding to five upper and four lower teeth, characteristic of both extant and extinct sloths (Delsuc et al. 2019). This evidence suggests continuity between sloths, meaning that they may form a single holobaramin.
Cingulata and the placement of Dasypus
There appear to be two cingulate holobaramins, the genus Dasypus and all other cingulates. It is rather interesting that the 13-foot-long extinct Doedicurus should be the closest to these two species of four-inch fairy armadillos. This shows the overall similarity between large, extinct armadillos and extant small-sized ones. However, this assertion is based only on mtDNA sequence similarity without taking the nuclear genome into account. Apparently, size reduction has also happened among the non-Dasypus armadillos, as has likely happened among the cat and dog kinds.
Delsuc at el. (2001) analyzed the mitochondrial 12S and 16S rRNA and the nuclear 28th exon of the vWF genes in eight xenarthran genera. According to their results, when examining the vWF alone, the grouping of Cabassous and Chaetophractus to the exclusion of Dasypus was mixed. However, when all three genes were combined, support for the exclusion of Dasypus increased. Delsuc et al. (2002) also found similar results when analyzing the concatenation of three nuclear genes: α2B adrenergic receptor (ADRA2B), the breast cancer susceptibility (BRCA1) gene, and the vWF for 12 of the 13 living xenarthran genera. Presslee et al. (2019) aligned 120 type 1 collagen protein sequences from 24 genus-level taxa. They found that the nine-banded armadillo (Dasypus novemcinctus) separated significantly from all other armadillo species, together with Doedicurus and Glyptodon.
Furthermore, Billet et al. (2011) analyzed 125 cranial characters from 22 armadillo taxa and found that the tolypeutine and euphractine armadillos, and glyptodontoids form a clade. However, they found dasypodines to be monophyletic but also less inclusive compared to the previously mentioned clade. This, as well as the molecular evidence from Delsuc et al. (2001; 2002), indicate that Dasypus may likely be a separate holobaramin.
Interestingly, D. novemcinctatus is the only known vertebrate species that exhibits obligate monozygotic polyembryony. During this process, a single blastocyst undergoes two cell divisions to produce genetically identical quadruplets of the same gender (Prodöhl et al. 1996). While there exists variation in size, scute number and patterns, this could well be due to non-genetic factors, such as cellular metabolite concentration or the number of mitochondria in each descendent cell. It is also well-known that after a time, even identical twins are not identical genetically. Considering this unique ontogenetic mode might indicate that Dasypus is a different holobarmin compared to other armadillos.
Conclusion and Outlook
As described in the Results and Discussion sections, there appear to be four xenarthran kinds: anteaters, sloths, and two armadillo groups. But how did xenarthrans get to the Americas from the Ark? This may be the subject of a subsequent paper, but some preliminary thoughts may be discussed here.
Until now, xenarthran fossils have only been discovered in South America, the Caribbean, and Central America. This distribution can be attributed to an event called the Great American Biotic Interchange (GABI) (Carrillo et al. 2015; Woodburne 2010). However, of note is the lack of xenarthran fossils in Africa. One would imagine that if animals wandered from the Ark all the way to the Americas, they would leave fossils at least somewhere along the way. This might not be such a big problem for the following reasons.
When the animals left the Ark, they would have initially formed smaller groups on their way to South America. Due to their sparse numbers, it is possible that many different vertebrate groups simply did not leave fossils along the way. Also remember that fossils usually do not form under normal conditions like what we observe today. Rather fossils must form quickly under special, catastrophic conditions.
According to a theory by de Queiroz (2014), some animals could have rafted across the ocean from Africa to South America shortly after the Flood (when the two continents were still close), just as animals could have rafted from mainland South America to the Galapagos islands. One such species is Ucayalipithecus perdita, a non-platyrrhine species of monkey from the family Parapithecidae. This same family of primates also exists in northern Africa, but fossils have also been found from this species in the Peruvian Andes (Seiffert et al. 2020).
This work uses morphological and genetic datasets to derive putative xenarthram baramins. This highlights the usefulness of additive evidence to classify species into separate holobaramins. With more genetic datasets and biogeographic data, we may get a more complete picture of the baraminology of anteaters, armadillos, and sloths.
Acknowledgments
The author would like to thank Matthew McLain for helping in the interpretation of the mitochondrial DNA analysis. Jim Brenneman also helped in the biblical analysis of xenarthrans based on Genesis 1:24–26.
References
Anderson, Robert P., and C. O. J. Handley. 2001. “A New Species of Three-Toed Sloth (Mammalia: Xenarthra) from Panamá, with a Review of the Genus Bradypus.” Proceedings of the Biological Society of Washington 114 (April): 1–33.
Billet, Guillaume, Lionel Hautier, Christian de Muizon, and Xavier Valentin. 2011. “Oldest Cingulate Skulls Provide Congruence Between Morphological and Molecular Scenarios of Armadillo Evolution.” Proceedings of the Royal Society B Biological Sciences 278, no. 1719 (22 September): 2791–2797.
Carrillo, Juan D., Analia Forasiepi, Carlos Jaramillo, and Marcelo R. Sánchez-Villagra. 2015. “Neotropical Mammal Diversity and the Great American Biotic Interchange: Spatial and Temporal Variation in South America’s Fossil Record.” Frontiers in Genetics (5 January) 5:451.
Chen, Vincent B., Ian W. Davis, and David C. Richardson. 2009. “KING (Kinemage, Next Generation): A Versatile Interactive Molecular and Scientific Visualization Program.” Protein Science 18, no. 11 (September 18): 2403–2409.
Cserhati, Matthew. 2020. “Do Lizards and Snakes Form Separate Apobaramins?” Journal of Creation 34, no. 1 (April): 96–101.
Cserhati, Matthew. 2021. “Molecular and Morphological Analysis Predicts Four Bat Baramins.” Journal of Creation 35, no. 1 (April): 117–127.
Cserhati, M. 2023. “Baraminic Placement of Homo heidelbergensis Based on Molecular Data.” Creation Research Society Quarterly 59, no. 3: 151–159.
Delsuc, F., F. M. Catzeflis, M. J. Stanhope, and E. J. Douzery. 2001. “The Evolution of Armadillos, Anteaters and Sloths Depicted by Nuclear and Mitochondrial Phylogenies: Implications for the Status of the Enigmatic Fossil Eurotamandua.” Proceedings of the Royal Society B, Biological Sciences 268, no. 1476 (August): 1605–1615.
Delsuc, Frédéric, Mark Scally, Ole Madsen, Michael J. Stanhope, Wilfried W. de Jong, Françis M. Catzeflis, Mark S. Springer, and Emmanuel J. Douzery. 2002. “Molecular Phylogeny of Living Xenarthrans and the Impact of Character and Taxon Sampling on the Placental Tree Rooting.” Molecular Biology and Evolution 19, no. 10 (October): 1656–1671.
Delsuc, Frédéric, Michael J. Stanhope, and Emmanuel J. P. Douzery. 2003. “Molecular Systematics of Armadillos (Xenarthra, Dasypodidae): Contribution of Maximum Likelihood and Bayesian Analyses of Mitochondrial and Nuclear Genes.” Molecular Phylogenetics and Evolution 28, no. 2 (August): 261–275.
Delsuc, Frédéric, Mariella Superina, Marie-Ka Tilak, Emmanuel J. P. Douzery, and Alexandre Hassanin. 2012. “Molecular Phylogenetics Unveils the Ancient Evolutionary Origins of the Enigmatic Fairy Armadillos.” Molecular Phylogenetics and Evolution 62, no. 2 (February): 673–680.
Delsuc, Frédéric, Gillian C. Gibb, Melanie Kuch, Guillaume Billet, Lionel Hautier, John Southon, Jean-Marie Rouillard, Juan Carlos Fernicola, Sergio F. Vizcaíno, Ross D. MacPhee, and Hendrik N. Poinar. 2016. “The Phylogenetic Affinities of the Extinct Glyptodonts.” Current Biology 26, no. 4 (22 February): R155–6. doi: 10.1016/j.cub.2016.01.039.
Delsuc, Frédéric, Melanie Kuch, Gillian C. Gibb, Emil Karpinski, Dirk Hackenberger, Paul Szpak, Jorge G. Martínez, Jim I. Mead, H. Gregory McDonald, Ross D. E. MacPhee, Guillaume Billet, Lionel Hautier, and Hendrik N. Poinar. 2019. “Ancient Mitogenomes Reveal the Evolutionary History and Biogeography of Sloths.” Current Biology 29, no. 12 (17 June): 2031–2042.
De Queiroz, Alan. 2014 The Monkey’s Voyage: How Improbable Journeys Shaped the History of Life. New York, NY: Basic Books.
Gaudin, Timothy J. 1995. “The Ear Region of Edentates and the Phylogeny of the Tardigrada (Mammalia, Xenarthra).” Journal of Vertebrate Paleontology 15, no. 3: 672–705.
Gaudin, Timothy J. 2004. “Phylogenetic Relationships Among Sloths (Mammalia, Xenarthra, Tardigrada): The Craniodental Evidence.” Zoological Journal of the Linnean Society 140, no. 2 (February): 255–305.
Gaudin, Timothy J., and Darin A. Croft. 2015. “Paleogene Xenarthra and the Evolution of South American Mammals.” Journal of Mammology 96(4): 622–34.
Gibb, Gillian C., Fabien L. Condamine, Melanie Kuch, Jacob Enk, Nadia Moraes-Barros, Mariella Superina, Hendrik N. Poinar, and Frédéric Delsuc. 2015. “Shotgun Mitogenomics Provides a Reference Phylogenetic Framework and Timescale for Living Xenarthrans.” Molecular Biology and Evolution 33, no. 3 (March): 621–642.
Mitchell, Kieren J., Agustin Scanferla, Esteban Soibelzon, Ricardo Bonini, Javier Ochoa, and Alan Cooper. 2016. “Ancient DNA from the Extinct South American Giant Glyptodont Doedicurus sp. (Xenarthra: Glyptodontidae) Reveals that Glyptodonts Evolved from Eocene Armadillos.” Molecular Ecology 25, no. 14 (July): 3499–3508.
Möller-Krull, Maren, Frédéric Delsuc, Gennady Churakov, Claudia Marker, Mariella Superina, Jürgen Brosius, Emmanuel J. P. Douzery, and Jürgen Schmitz. 2007. “Retroposed Elements and Their Flanking Regions Resolve the Evolutionary History of Xenarthran Mammals (Armadillos, Anteaters, and Sloths).” Molecular Biology and Evolution 24, no. 11 (November): 2573–2582.
Novacek, Michael J. 1992. “Mammalian Phylogeny: Shaking the Tree.” Nature 356, no. 6365 (April): 121–125.
Oliver, Jillian D., Katrina E. Jones, Lionel Hautier, W. J. Loughry, and Stephanie E. Pierce. 2016. “Vertebral Bending Mechanics and Xenarthrous Morphology in the Nine-Banded Armadillo (Dasypus novemcinctus).” Journal of Experimental Biology 219, no. 19 (October): 2991–3002.
Poinar, Hendrik, Melanie Kuch, Gregory McDonald, Paul Martin, and Svante Pääbo. 2003. “Nuclear Gene Sequences from a Late Pleistocene Sloth Coprolite.” Current Biology 13, no. 13 (July 1): 1150–1152.
Presslee Samantha, Graham J. Slater, François Pujos, Analía M. Forasiepi, Roman Fischer, Kelly Molloy, Meaghan Mackie, et al. 2019. “Palaeoproteomics Resolves Sloth Relationships.” Nature Ecology and Evolution 3 (July): 1121–1130.
Prodöhl, P. A., W. J. Loughry, C. M. McDonough, W. S. Nelson, and J. C. Avise. 1996. “Molecular Documentation of Polyembryony and the Micro-Spatial Dispersion of Clonal Sibships in the Nine-Banded Armadillo, Dasypus novemcinctus.” Proceedings of the Royal Society B: Biological Sciences 263, no. 1377 (December 22): 1643–1649.
Seiffert, Erik R., Marcelo F. Tejedor, John G. Fleagle, Nelson M. Novo, Fanny M. Cornejo, Mariano Bond, Dorien de Vries, and Kenneth E. Campbell Jr. 2020. “A Parapithecid Stem Anthropoid of African Origin in the Paleogene of South America.” Science 368, no. 6487 (10 April): 194–197.
Slater, Graham J., Pin Cui, Analía M. Forasiepi, Dorina Lenz, Kyriakos Tsangaras, Bryson Voirin, Nadia de Moraes-Barros, Ross D. E. MacPhee, and Alex D. Greenwood. 2016. “Evolutionary Relationships among Extinct and Extant Sloths: The Evidence of Mitogenomes and Retroviruses.” Genome Biology and Evolution 8, no. 3 (14 February): 607–621.
Wood, T.C. 2020. “Expanding the Toolkit of Statistical Baraminology with BARCLAY: Baraminology and Cluster Analysis.” Journal of Creation Theology and Science Series B: Life Sciences 10:7.
Woodburne, Michael O. 2010. “The Great American Biotic Interchange: Dispersals, Tectonics, Climate, Sea Level and Holding Pens.” Journal of Mammalian Evolution 17, no. 4 (July 14)): 245–264.