The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
Previous plant molecular baraminology studies included analysis of the chloroplast genome alone (Liliales), as well as comparison to hybridization data (Cucurbitaceae). The present study includes the analysis of chloroplast genomes and mitochondrial genomes in various species of myrtles (Myrtales).
Myrtales is an economically important order of rosids comprising nine families, 380 genera, and approximately 13,000 species. Compared to other angiosperms with a single phloem layer, Myrtales uniquely possesses phloem tissue on both sides of its xylem vessels.
The chloroplast genomes of 306 Myrtales and six Cucurbita species were downloaded from NCBI and aligned using the MAFFT sequence alignment software. Using ward.D2 clustering, 15 statistically significant clusters were found (p<0.05), with at least three species (Melastoma, Syzygium, Angophora + Corymbia + Eucalyptus, Phyllagathis, Lagerstroemia, Sonneratia, Sonerila, Trapa, Circaea + Ludwigia, Chamaenerion, Epilobium, Oenothera, and four mixed groups). Oenothera curtiflora, lindheimeri, and villaricae contained a genomic inversion encompassing the genes rbcL, atpB, atpE, rps4, psaA–D, petN, rpoB, ropC1, rpoC2, rps2, atpI, atpH, atpF, atpA, psbI, and psbK. This inversion likely occurred before seven other Oenothera species split off and formed a monobaramin.
The cytochrome-c oxidase subunit I (COI) gene sequence was extracted for 18 Myrtales species having mitochondrial genomes, with Brassica napus (rapeseed) as an outlier. These sequences were aligned and clustered. Three statistically significant clusters were identified, including the genera Oenothera (O. biennis, elata and villaricae), Eucalyptus, and Melastoma. These three genera were also found by the chloroplast analysis, meaning that with two separate lines of evidence, these may indeed be real plant holobaramins.
Keywords: Myrtales, chloroplast, mitochondrion, baraminology, Oenothera, Syzygium, Lagerstroemia, Eucalyptus, Trapa, Epilobium
Introduction
Myrtales (myrtles) is an order of angiosperms comprising nine families, 380 genera, and more than 13,000 species, mostly trees and shrubs (Zhang et al., 2021). Myrtales is a basal rosid in the group of core eudicots classified in the Angiosperm Phylogeny Group IV (APG IV). The nine families in the order consist of Alzateacaea, Combretaceae, Crypteroniaceae, Lythraceae, Melastomataceae, Myrtaceae, Onagraceae, Peneaceae, and Vochysiaceae. The species richness of these families is uneven, however, with the majority of species in Myrtales belonging to Myrtaceae and Melastomataceae. Most members of the Myrtales group are found in tropical and subtropical environments and notably include Eucalyptus trees, the classic myrtle, pomegranate, and other food, spice, and ornamental plants. Several Myrtales species have economic importance in the production of essential oils (Dillmann et al. 2020) and their secondary metabolites also have medicinal uses (Maphetu et al. 2022).
Members of Myrtales are primarily distributed in warmer regions with almost no representation in colder climates, although some species are found in Tasmania and Southern Australia. The two biggest families, Myrtaceae and Melastomataceae, are predominantly found in Australia and in the New World. Myrtaceae, consisting of 131 genera and more than 5,900 species, is the largest family in Myrtales. While most of its members found in Australia are dry-fruited, its American representatives mostly yield berries. Melastomataceae is the second-largest family, with 188 genera and more than 4,960 species. More than two-thirds of Melastomataceae are found in the New World, consisting largely of shrubs and small trees.
Most angiosperms possess only one phloem layer. However, members of Myrtales are unique in possessing phloem tissue on both sides of their xylem vessels. Plants typically contain structures called pits that line the vessel walls in stems. In Myrtales, these pits have a sieve-like appearance, called vestured pits. These features of wood anatomy are otherwise rarely observed in flowering plants and help lend morphological definition to the order.
The goal of this study is to analyze the order of Myrtales from a molecular baraminological aspect. Baraminology is the study of the biblical kinds mentioned in Genesis 1: 11–12, 21, 24–25. The kinds of animals and plants mentioned in these verses are reproductive communities that can interbreed with one another but don’t interbreed with species from other kinds. A biblical kind is called a ‘holobaramin’, to use a technical term. Holobaramins correspond largely to the level of family.
To do this we use chloroplast genomes from this order to classify members of Myrtales into several putative holobaramins. Chloroplasts are abundant in plant cells, and because of their short size, they are easy to isolate and sequence. Furthermore, their gene repertoire, gene order, and sequence similarity are conserved (Wicke et al. 2011). This makes the chloroplast genome an ideal tool to measure species similarity. This way we can classify two species with high sequence similarity to the same baramin whereas species with low sequence similarity belong to different baramins. The caveat is that due to their small size (150 Kbp), the chloroplast genome may not be representative of the entire genome, furthermore, since it is maternally inherited, it may only represent the maternal line.
Materials and Methods
Sequences
We downloaded 306 Myrtales RefSeq chloroplast genomes and 37 Myrtales mitochondrial genomes from NCBI. Ten chloroplast genomes were removed because they were redundant, and eight were removed, because the start of their sequence was very different than the great majority of sequences. This left 288 genomes. We also downloaded seven Cucurbita chloroplast genomes and the mitochondrion genome for Brassica napus (rapeseed, order Brassicales) from NCBI’s online database as outgroups, that is, a clearly distinct kind to function as a control group (see Supplementary Files 1 and 2).
Alignment software and parameters
We used the MAFFT 7.520 alignment software (Katoh and Standley 2013) for Windows (https://mafft.cbrc.jp/alignment/software/windows_without_cygwin.html) to align the chloroplast genomes of each species. We used the default parameters, with the output file being an unsorted fasta alignment file. Running MAFFT on entire mitochondrial genome sequences was very slow, due to the average mitochondrial genome being about twice as long as the average chloroplast genome. Instead, the gene sequence for cytochrome oxidase subunit I (COI) was extracted from 18 of the 37 mitochondrial genomes. 18 of the mitochondrial genomes were excluded because they were from the same species (most likely different strains in the same species). The B. napus COI gene (extracted from NC_008285.1) was BLASTED against the mitogenome for three of these 18 species. These COI gene sequences were then aligned using CLUSTALW (Hung and Weng 2016). Then the sequence identity matrix was defined using our own R script using the ‘dist.alignment’ command from the ‘seqinr’ package (setting the matrix parameter to identity).
Figures and tables
The genome length and GC% image in fig. 1 was created using the ‘ggplot2’ R package. The ‘heatmap’ command using the ward.D2 method was used to create the heatmaps in figs. 2 and 3. The OGDRAW online software (Greiner, Lehwark, and Bock 2019) was used to draw the chloroplast genome maps of several Oenothera species in fig. 4. The effect sizes in table 1 were calculated using the ‘effectsize’ package in R.
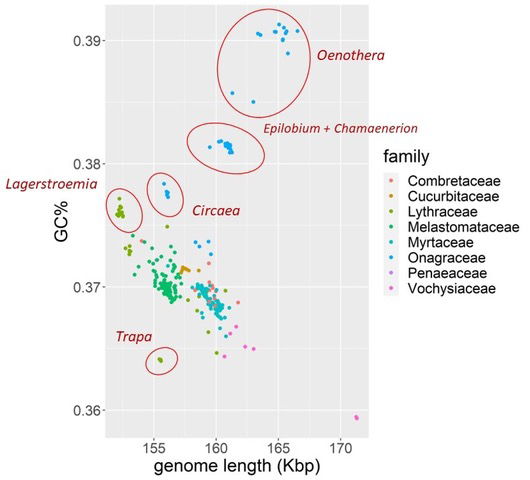
Fig. 1. All of the Myrtales species in this study plotted based on their chloroplast genome length and the GC% of their genome. Species are plotted based on which family they belong to. Five groups separate from all the other species: Oenothera, Epilobium + Chamaenerion, Circaea, Lagerstroemia, and Trapa.
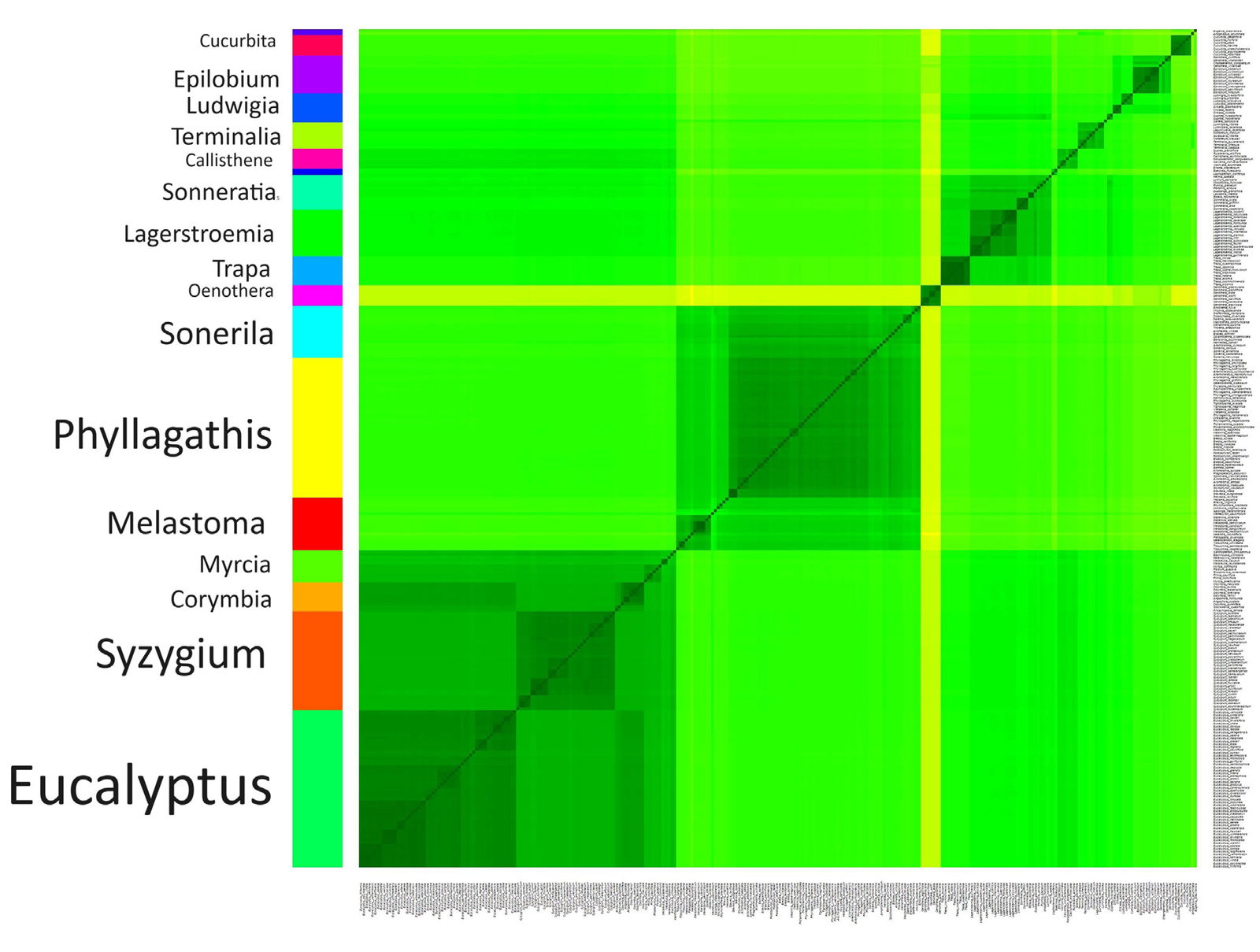
Fig. 2. Heatmap depicting the 18 putative clusters found in the chloroplast genome analysis. The 288 species were clustered according to genome sequence similarity. Darker green colors correspond to higher sequence similarity, meaning that these species likely belong to the same baramin. Lighter, yellower colors correspond to lower sequence similarity, meaning that these species likely belong to different baramins.
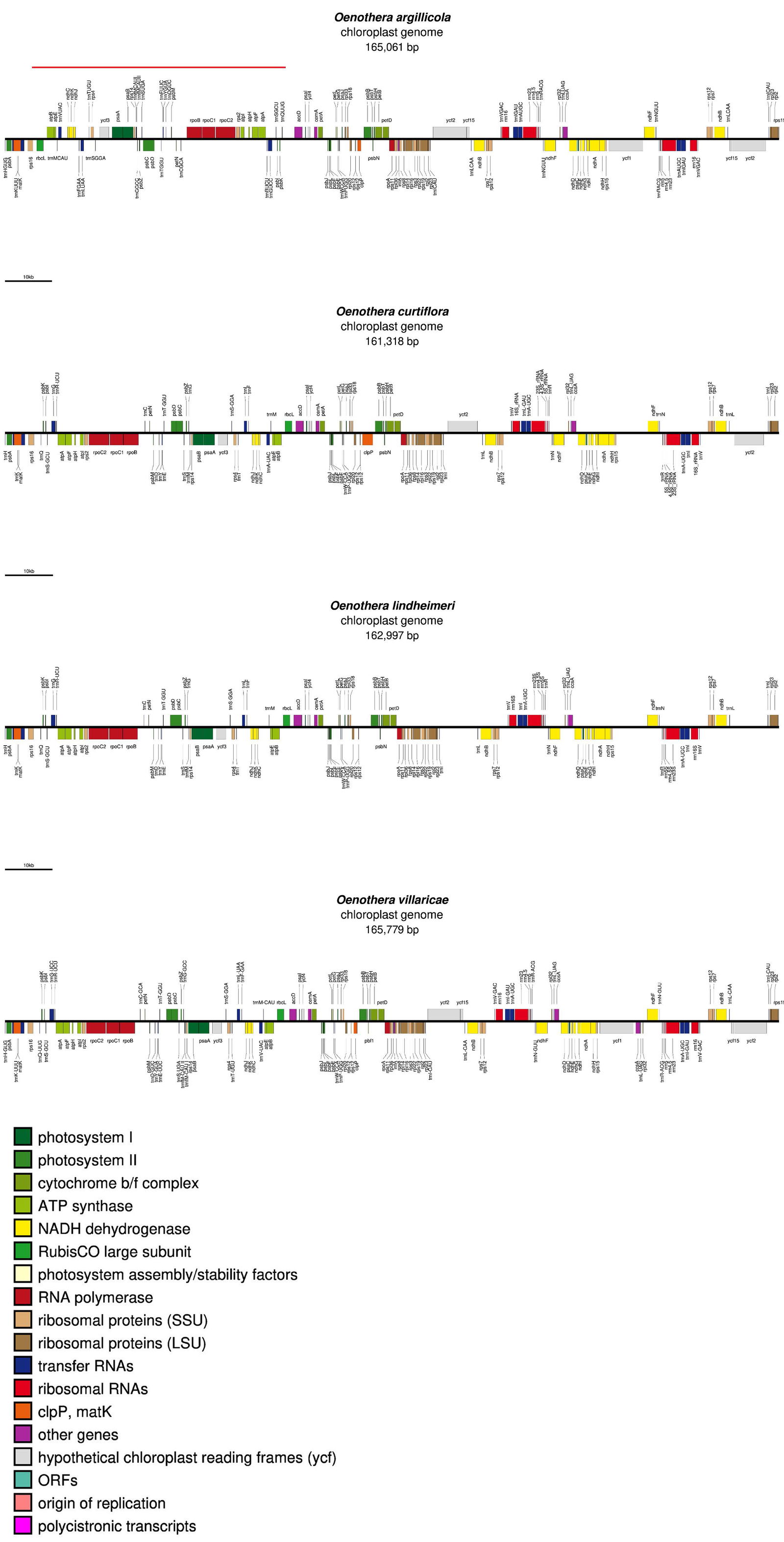
Fig. 3. Chloroplast genome maps of Oenothera argillicola, curtiflora, lindheimeri, and villaricae. The red line denotes the genome inversion that happened between the latter three species and all other species of Oenothera.
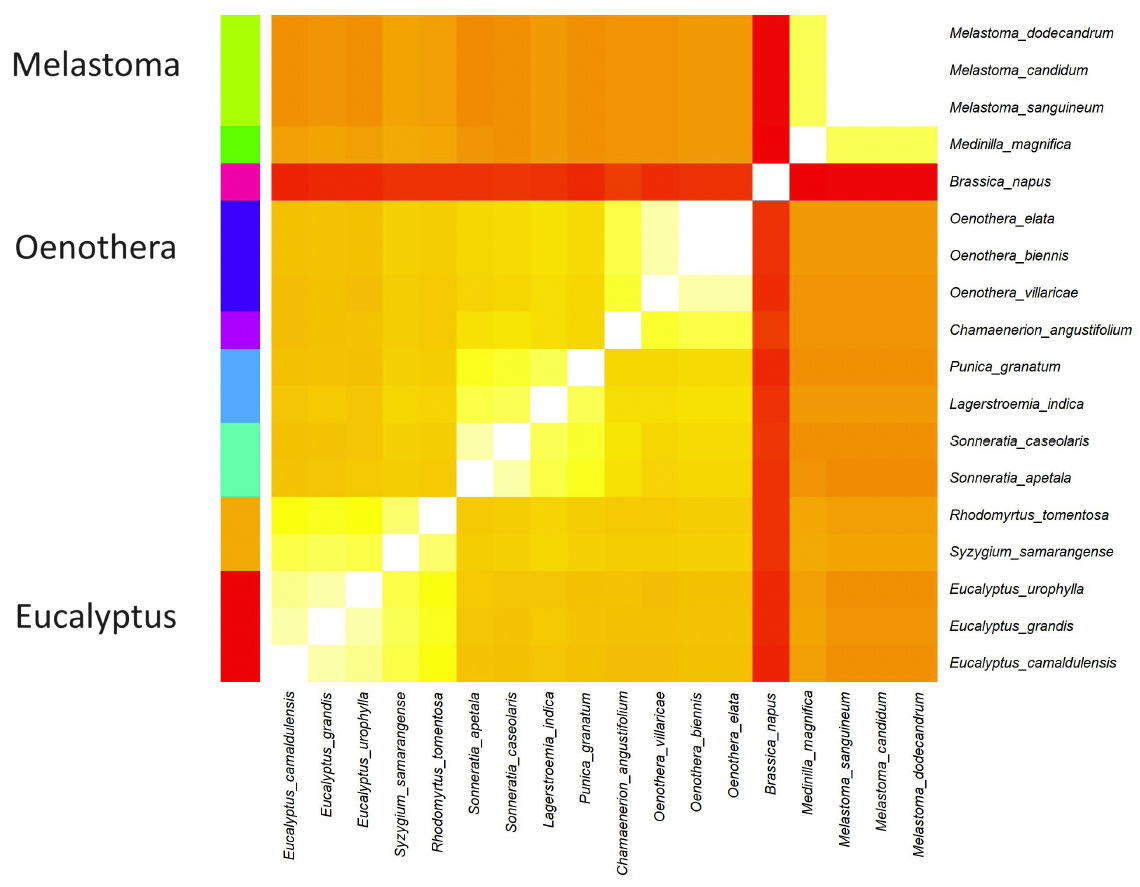
Fig. 4. Heatmap depicting the nine putative clusters found in the COI gene analysis. The 18 species were clustered according to genome sequence similarity. Lighter yellower colors correspond to higher sequence similarity, meaning that these species likely belong to the same baramin. Darker, redder colors correspond to lower sequence similarity, meaning that these species likely belong to different baramins.
Availability
Supplementary Files 1 and 2 are available on Zenodo at https://zenodo.org/records/10729431.
Biblical analysis
As plants, myrtles were created on Day 3 of Creation week, separate from all other organisms. Of all the myrtle species, the Bible mentions myrtles (genus Myrtus) five times, and the pomegranate (Punica granatum) 12 times as actual plants and not ornaments. Zechariah 1:8, 10–11 mentions myrtle trees by themselves. However, Nehemiah 8:15, Isaiah 41:19, and Isaiah 55:13 each mention the myrtle tree besides other species of trees. For example, Nehemiah 8:15 mentions oil trees, olive branches, myrtles, and palm branches. However, the “branches of leafy trees” are also mentioned. This could mean that the olive tree, the myrtle and palm trees belong to separate kinds of leafy trees. However, this does not shed much light onto baraminic relationships within myrtles.
1 Samuel 14:2 mentions an individual pomegranate tree. Numbers 13:23, 20:5, Deuteronomy 8:8, Song of Solomon 6:11, 7:12, Joel 1:12, and Haggai 2:19 mention pomegranates besides the olives tree, the apple tree, figs, wheat, barley and vines. Thus, we could reason that pomegranates are at least a separate species within myrtles, which are distinct from olives, apples, figs, wheat, barley and vines. Song of Solomon 4:3 and 6:7 only describe the physical qualities, such as the texture of the pomegranate.
Not much can be inferred from the Bible about myrtles. Myrtles apparently are an apobaramin, which is distinct from other groups of plants, such as palms, olives, or figs. However, the pomegranate appears to belong to its own group, separate from other myrtle species.
Results and Discussion
Chloroplast genome length and GC%
On a large-scale level the chloroplast genomes were compared based on their genome length and their GC%. If species belong to the same holobaramin, we may expect that their gene repertoire, gene order, genome length, sequence similarity and GC% are all conserved. The results can be seen in fig. 1. Three groups from the family Onagraceae and one from Lythraceae seem to form clusters, separate from all the other species. Surprisingly, three dots can be seen representing Oenothera curtiflora, O. lindheimeri, and O. villaricae between the main Oenothera group and Epilobium + Chamaenerion.
One group, with most species having a genome length between 162.5–167.5 Kbp and a GC% around 39% corresponds to the genus Oenothera.
Another group, with a clp genome less than 152.5 Kbp and a GC% between 37.5% and 38% corresponds to the 17 species from the genus Lagerstroemia from the family Lythraceae. Yet another group of five species with a GC% between 37.5% and 38% with a genome length between 155 Kbp and 157.5 Kbp corresponds to the genus Circaea (family Onagraceae).
Fifteen other species belonging to the genera Epilobium and Chamaenerion (family Onagraceae) clustered together with a GC% between 38% and 38.5%, and a genome size between 160 and 162.5 Kbp, with an outlier, Ch. conspersum, with a genome size of 159,496 bp.
In a study of 13 Circaea, Chamaenerion and Epilobium species by Luo et al. (2021), the genome structure of Chamaenerion and Epilobium were found to be similar to one another, and different from those of Circaea. The latter genus had a larger variety of repeat sequences compared to the other two.
Ten species of Trapa form their own group with a clp genome length of around 155 Kbp and a GC% of approximately 36.5%.
Chloroplast genome sequence analysis
The analysis of the chloroplast genomes found 15 putative statistically significant clusters (p < 5%) with at least three members. The Hopkins clustering statistic was 0.965, which means outstanding clustering. In the following, these 15 putative Myrtales holobaramins will be described.
The first cluster with 18 species is dominated by species from the genera Melastoma, Osbeckia, and Tibouchina. Therefore, this cluster was called the Melastoma group. Two member genera, Melastoma and Osbeckia, have been found to contain the same trichromal whole genome duplication, supporting their holobaraminic status (Zhong et al. 2023). There is additional strong evidence of hybridization between members of the Melastoma genus (Liu et al. 2014).
The second group consists of 34 species from the genus Syzygium from the family Myrtaceae. This family is very diverse with approximately 1,200 species across Africa and Asia. Myrtaceae has four large genera, one of them which is Syzygium (Maurin et al. 2021). Infrageneric classifications of Syzygium have been conducted, but the genus is not well studied (Craven and Biffen 2010).
Group three is made up of ten species mainly from the genus Corymbia (bloodwood eucalypts, six species) and two from Angophora, both from Myrtaceae, tribe Eucalypteae. Included in this group is the monotypic genus Stockwellia, whose only member species, Stockwellia quadrifida, is endemic to Queensland (Carr et al. 2002). These two genera classify as eucalypts, or gum trees, besides the genus Eucalyptus (group #8). Together, these three genera (Angophora, Corymbia, and Eucalyptus) may possibly classify as the eucalypt apobaramin, since they are separate, however, this is not so clear, since the sequence similarity values between Corymbia and Eucalyptus are relatively high.
The eighth group consists of 54 species from the genus Eucalyptus (family Myrtaceae), the largest cluster found in this study, with a mean sequence similarity of 0.945. Eucalyptus is one of four large genera from the family Myrtaceae (Maurin et al. 2021). Of the 54 species, Eu. globulus belongs to the subgenus Symphyomyrtus, whereas all the others belong to the subgenus Monocalyptus.
Bayly (2016) considers Eucalyptus as the sister group of Corymbia + Angophora. It could be that all eucalypts could possibly be placed into a single holobaramin. According to Table 1A, when we compare all sequence similarity values between the two putative holobaramins Eucalyptus and Corymbia, we see that the mean sequence similarity is 0.897 ± 0.005. When we compare these similarity values to the similarity values within Eucalyptus (Table 1B, row 1), we see that the effect size is very small, only 0.047. There is also a very small effect size between Corymbia and Corymbia versus Eucalyptus (0.032, row #2, Table 1B). When we compare sequence similarity values between Eucalyptus and all non-Eucalyptus species, the effect size is 0.202. The effect size between Corymbia and all non-Corymbia species is 0.162. These differences constitute a small, yet discernible difference. This probably means that the differences between the holobaramins Eucalyptus and Corymbia are negligible to the point that these two groups could be united into a single eucalypt holobaramin.
Table 1. Sequence similarity and effect size between Eucalyptus and Corymbia
| A. Sequence similarity | |
|---|---|
| Eucalyptus vs. Eucalyptus | 0.945 ± 0.018 |
| Corymbia vs. Corymbia | 0.929 ± 0.032 |
| Eucalyptus vs. Corymbia | 0.897 ± 0.005 |
| Eucalyptus vs. all other species | 0.743 ± 0.079 |
| Eucalyptus vs. all other species | 0.768 ± 0.092 |
| B. Effect size | |
| Eucalyptus vs. (Eucalyptus vs. Corymbia) | 0.047 |
| Corymbia vs. (Eucalyptus vs. Corymbia) | 0.032 |
| Eucalyptus vs. all other species | 0.202 |
| Corymbia vs. all other species | 0.162 |
The next group, #4 consists of 48 species, and is the second largest cluster in our results. The genera, Allomorphia, Blastus, Bredia, Fordiophyton, Gravesia, Medinilla, and Phyllagathis (family Melastomataceae) all have at least three species in this group. When Zhou et al. (2020) analyzed the internal transcribed spacer (ITS), four nuclear genes (Dbr1, SOS4a, SOS4b, and PCRF1), and the intergenic spacer trnV-trnM, and found differentially fixed nucleotide substitutions in the hybrid between Phyllagathis longicalcarata and Sporoxeia petelotii (the latter species coming from one of the genera in cluster #4). Clausing and Renner (2001) found Medinilla, Gravesia, Driessenia, Phyllagathis, and Blastus to be all in the same clade based on their analysis of the genes rbcL, ndhF, and rpl16.
Group #5 consists of nine species from various genera, from the family Combretaceae. Member genera include Combretum, Lumnitzera, Laguncularia, Quisqualis, and Terminalia. There is molecular evidence that two species represented in this study, Lumnitzera littorea and Lumnitzera racemosa, are capable of natural hybridization, providing additional additive evidence (Guo et al. 2011).
Eleven species belong to group #6, which also consists of various genera. All genera belong to the family Mertaceae. Important member genera include Myrcia, Backhousia, Heteropyxis, Melaleuca, and Plinia. Phylogenetic research has pointed to the genus Myrcia being non-monophyletic, which supports its monobaraminic status as identified in this study (Lima et al. 2021).
Group #7 is made up of 16 species from the genus Lagerstroemia, from the family Lythraceae. This group was also found to be a separate group based on the analysis of GC% and clp genome length, which supports the idea that this genus is indeed a separate holobaramin. Hybridization of some species of Lagerstroemia has been observed, supporting its holobaraminic status (Pounders, Rinhart, and Sakhanokho 2007).
Group #9 consists of 12 species from various genera, four of which come from the genus Sonneratia. These species come from the family Lythraceae. The Sonneratia genus, which consists of six species, is known to readily hybridize intragenerically (Xie et al. 2020). The tenth cluster consists of 18 species in 15 genera. The species in this cluster belong to the family Melastomataceae.
Group #10 consists of 18 species, also from various groups, from the family Melastomataceae. Four species come from the genus Sonerila.
Group #11 consists of ten species from the genus Trapa, which belongs to the family Lythraceae. These species separate from all other species based on their GC% of around 36.5% and their clp genome length of around 155 Kbp. The tight clustering of the Trapa group is reflected by its exceptionally high sequence similarity index.
Ten species from four genera belong to group #12, namely Circaea, Cuphea, Ludwigia, and Saltera. Notably, these genera come from three different families: Oenograceae, Lythraceae, and Penaeaceae.
Group #13 contains only two species from Myrtaceae, Baeckea frutescens, and Lophostemon confertus. Group #14 also consists of two species only, Anogeissus acuminata and Eugenia brasiliensis. Neither of these two small groups were statistically significant.
Group #15 is rather interesting, as it is made up mainly of 13 species, nine of which come from the genus Epilobium, as well as Chamaenerion conspersum and three species of Oenothera (O. curtiflora, O. lindheimeri, and O. villaricae), all from the family Onagraceae. In contrast, seven other species of Oenothera belong to group #16. However, as we can see in fig. 1, all Oenothera species cluster together based on clp genome length and GC%. Chamaenerion is often included within Epilobium, supporting their inclusion in the same group (Wagner, Hoch, and Raven 2007).
The reason for this separation may be due to an approximately 56 Kbp inversion between the gene rbcL and the tRNA trnQ-UUG (see red line in fig. 3 above O. argillicola). Furthermore, two introns were found to be missing from the clpP gene from the larger group of Oenothera species from group #16, which were left intact in the smaller group of three species from group #15. These two introns typically occur in angiosperm species (Luo et al. 2021). The loss of these two introns and the 56 Kbp inversion could have led to the formation of a monobaramin within the genus Oenothera, with O. curtiflora, O. lindheimeri, and O. villaricae as members closer to the Oenothera archebaramin. This also explains why these three species are somewhat separated from the main Oenothera group in fig. 1.
Interestingly, a similar inversion event occurred in the clp genome of three species of Veratrum in the order Liliales (Cserhati 2023). Apparently, such large-scale inversions can occur within the clp genome in plants and may affect the calculation of sequence similarity within members of a holobaramin.
Group #17 consists of seven species, each from a separate genus of the family Vochysiaceae: Callisthene erythroclada, Erisma bracteosum, Korupodendron songweanum, Qualea grandiflora, Ruizterania albiflora, Salvertia convallariodora, and Vochysia acuminata. Previous research has found mixed support for the monophyletic status of Vochysiaceae rendering this baraminological result ambiguous (Maurin et al. 2021). Represented in this group is Korupodendron, a monotypic genus found only in West Central Africa (Litt and Cheek 2002).
Group #18 consists of the seven Cucurbita outlier species.
Mitochondrion genome sequence analysis
The 18 Myrtales species which had a COI sequence were aligned, and the sequence similarity matrix (see Supplementary File 2) was visualized in a heatmap in fig. 4. The Hopkins clustering statistic was 0.804 which corresponds to good clustering. Overall, nine clusters were found. Of these, three statistically significant (p < 5%) groups have at least three species. These are Eucalyptus, Melastoma, and Oenothera.
These results support the inferences that we can draw from the analysis of the clp genomes, namely that Eucalyptus forms a single holobaramin, and that Melastoma also belong to a larger holobaramin. This is additive evidence from both the chloroplast and the mitochondrial genome analysis that the clusters that we found correspond to real holobaramins.
Interestingly, the analysis of COI in Oenothera shows that O. villaricae and O. elata belong to the same group, which is the same thing as what we see in fig. 1. This supports the idea that all Oenothera species form a single holobaramin, despite how the chloroplast genome sequence analysis divides this genus into two groups.
Conclusion
The present study is the first to combine data from clp genomes as well as mitochondrial genes. Overall, 15 putative statistically significant clusters of species were found in the chloroplast analysis. Of these, the most significant are Angophora + Corymbia + Eucalyptus, Phyllagathis, Syzygium, Melastoma, Sonerila, Lagerstroemia, Epilobium, and Oenothera. Melastoma, Eucalyptus, and Oenothera were validated by both studies. A significantly large inversion was discovered in the clp genome of seven species of Oenothera, which we could use to delineate a monobaramin. This is similar to an inversion found in the genus Veratrum in a previous clp genome baraminology study.
As more and more studies are performed based on chloroplast genomes using evidence from hybridization and mitochondrial data, we can be more confident in our predictions of holobaramins within plants. Adding hybridization data or data from nuclear genes as additive evidence would lend further support in verifying the holobaramins predicted in this study.
This is just one of the first chloroplast genome-based baraminology studies, with hopefully many more to follow. Chloroplast genome analysis can greatly help the baraminic assessment of plant species.
References
Bayly Michael J. 2016. “Phylogenetic Studies of Eucalypts: Fossils, Morphology and Genomes.” Proceedings of the Royal Society of Victoria 128, no. 1 (10 August): 12–24.
Carr, Denis J., Carr, S. G. M., B. P. M. Hyland, Peter G. Wilson, and Pauline Y. Ladiges. 2002. “Stockwellia quadrifida (Myrtaceae), a New Australian Genus and Species in the Eucalypt Group.” Botanical Journal of the Linnean Society 139, no. 4 (August): 415–421.
Clausing G., and S. S. Renner. 2001. “Molecular Phylogenetics of Melastomataceae and Memecylaceae: Implications for Character Evolution.” American Journal of Botany 88, no. 3 (March): 486–498.
Craven, L. A., and Ed Biffin. 2010. “An Infrageneric Classification of Syzygium (Myrtaceae).” Blumea—Biodiversity, Evolution and Biogeography of Plants 55, no. 1 (April): 94–99.
Cserhati, Matthew. 2023. “Chloroplast Genome-Based Baraminology Study of Liliales.” Creation Research Society Quarterly 60 (Fall): 84–96.
Dillmann, Janaina Brand, Luciana Filippin Cossetin, Marjorie de Giacometi, Dionatan Oliveira, Antônio Francisco Igor Magalhães de Matos, Pamela Daniele Avrella, Quelen Iane Garlet, Berta Maria Heinzmann, and Silvia Gonzalez Monteiro. 2020. “Adulticidal Activity of Melaleuca alternifolia (Myrtales: Myrtaceae) Essential Oil With High 1,8-Cineole Content Against Stable Flies (Diptera: Muscidae).” Journal of Economic Entomology 113, no. 4 (August): 1810–1815.
Greiner, Stephan, Pascal Lehwark, and Ralph Bock. 2019. “OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes.” Nucleic Acids Research 47, no. W1 (July 2): W59–W64.
Guo, Miaomiao, Renchao Zhou, Yelin Huang, Jianhua Ouyang, and Suhua Shi. 2011. “Molecular Confirmation of Natural Hybridization Between Lumnitzera racemosa and Lumnitzera littorea.” Aquatic Botany 95, no. 1 (July): 59–64.
Hung, Jui-Hung, and Zhiping Weng. 2016. “Sequence Alignment and Homology Search with BLAST and ClustalW.” Cold Spring Harbor Protocols 2016, no. 11 (November). doi:10.1101/pdb.top093070.
Katoh, K., and Standley, D. M. 2013. “MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability.” Molecular Biology and Evolution 30, no. 4 (April): 772–780.
Lima, Duane F., Renata Goldenberg, Félix Forest, Robyn S. Cowan, and Eva J. Lucas. 2021. “Phylogeny and Biogeography of Myrcia sect. Aguava (Myrtaceae, Myrteae) Based on Phylogenomic and Sanger Data Provide Evidence for a Cerrado Origin and Geographically Structured Clades.” Molecular Phylogenetics and Evolution 157 (April): 107043.
Litt, Amy, and Martin Cheek. 2002. “Korupodendron Songweanum, a New Genus and Species of Vochysiaceae from West-Central Africa.” Brittonia 54, no. 1 (January–March): 13–17.
Liu, Ting, Yunyun Chen, Lifang Chao, Shuqiong Wang, Wei Wu, Seping Dai, Feng Wang, Qiang Fan, and Renchao Zhou. 2014. “Extensive Hybridization and Introgression Between Melastoma candidum and M. sanguineum.” PLoS ONE 9, no. 5 (May 5): e96680. https://doi.org/10.1371/journal.pone.0096680.
Luo, Yike, Jian He, Rudan Lyu, Jiamin Xiao, Wenhe Li, Min Yao, Linying Pei, Jin Cheng, Jinyu Li, and Lei Xie. 2021. “Comparative Analysis of Complete Chloroplast Genomes of 13 Species in Epilobium, Circaea, and Chamaenerion and Insights Into Phylogenetic Relationships of Onagraceae.” Frontiers in Genetics 12 (4 November): 730495. https://doi.org/10.3389/fgene.2021.730495.
Maphetu, Nhlanhla, Jeremiah Oshiomame Unuofin, Nelisiwe Prenate Masuku, Chijioke Olisah, and Sogolo Lucky Lebelo. 2022. “Medicinal Uses, Pharmacological Activities, Phytochemistry, and the Molecular Mechanisms of Punica granatum L. (pomegranate) Plant Extracts: A Review.” Biomedicine and Pharmacotherapy 153 (September): 113256. https://doi.org/10.1016/j.biopha.2022.113256.
Maurin, Olivier, Artemis Anest, Sidonie Bellot, Edward Biffin, Grace Brewer, Tristan Charles-Dominique, Robyn S. Cowan, et al. 2021. “A Nuclear Phylogenomic Study of the Angiosperm Order Myrtales, Exploring the Potential and Limitations of the Universal Angiosperms353 Probe Set.” American Journal of Botany 108, no. 7 (July): 1087–1111.
Pounders, Cecil, Tim Rinehart, and Hamidou Sakhanokho. 2007. “Evaluation of Interspecific Hybrids between Lagerstroemia indica and L. speciosa.” HortScience 42, no. 6 (1 October): 1317–1322.
Wagner, Warren L., P. C. Hoch, and Peter H. Raven. 2007. “Revised Classification of the Onagraceae.” Systematic Botany Monographs 83: 1–240.
Wicke, Susann, Gerald M. Schneeweiss, Claude W. dePamphilis, Kai F. Müller, and Dietmar Quandt. 2011. “The Evolution of the Plastid Chromosome in Land Plants: Gene Content, Gene Order, Gene Function.” Plant Molecular Biology 76, nos. 3–5 (July): 273–297.
Xie, Wei, Cairong Zhong, Xinnian Li, Zixiao Guo, and Suhua Shi. 2020. “Hybridization with Natives Augments the Threats of Introduced Species in Sonneratia Mangroves.” Aquatic Botany 160 (January): 103166.
Zhang, Xiao-Feng, Jacob B. Landis, Hong-Xin Wang, Zhi-Xin Zhu, and Hua-Feng Wang. 2021. “Comparative Analysis of Chloroplast Genome Structure and Molecular Dating in Myrtales.” BMC Plant Biology 21: 219.
Zhong, Yan, Wie Wu, Chenyu Sun, Peishan Zou, Ying Liu, Seping Dai, and Renchao Zhou. 2023. “Chromosomal-Level Genome Assembly of Melastoma candidum Provides Insights into Trichome Evolution.” Frontiers in Plant Science 14 (27 January): 1126319. https://doi.org/10.3389/fpls.2023.1126319.
Zhou, Shuaixi, Shuheng Ni, Jinhong Dai, Qiujie Zhou, Renchao Zhou, and Ying Liu. 2020. “Natural Hybridization Between Phyllagathis and Sporoxeia Species Produces a Hybrid Without Reproductive Organs.” PLoS One 15, no. 1 (January 8): e0227625.
Supplementary Materials
Supplementary Files 1 and 2 are available on Zenodo at https://zenodo.org/records/10729431.





