The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
Amphibians are a class of vertebrates that are generally small, soft-skinned, and can live both on land and in water. They can also be either fully aquatic or fully terrestrial (Pough 2007). Three extant orders belong to Amphibia, namely Anura (frogs and toads), Caudata (salamanders and newts), and Gymnophiona (caecilians). Previous baraminology studies have identified 28 amphibian baramins.
The present study analyzed the mitochondrial genome of 362 amphibian species from the National Center for Biotechnology Information Database. C-values are also analyzed to estimate genome size using the Genome Size Analysis tool at The Molecular Baraminology Analysis Tool Suite (Cserhati and Maggs 2025). This was done to augment morphological results from previous studies with molecular results.
Furthermore, two new statistical tests were introduced to better filter putative holobaramins. These included skewness tests that measure how much sequence similarity values deviate from the normal distribution and the Dip test that measures unimodality.
Four putative anuran kinds were found which corresponded to the genera Bombina, Kaloula, Pelophylax, and Xenopus. Three putative caudatan kinds were also predicted: the genera Echinotriton + Tylototriton, Triturus, and Ambystoma. Lastly, two putative gymnophionan kinds were found: Caecilia + Oscaecilia (family Caeciliidae), Microcaecilia (family Siphonopidae), and one species from each of the genera Grandisonia, Hypogeophis, and Praslinia (family Grandisoniidae). However, the results from the analysis of the order Gymnophiona were not statistically significant enough to derive final conclusions. Therefore, it is possible that Gymnophiona consists of a single holobaramin.
Keywords: amphibian, Anura, Caudata, Gymnophiona, molecular baraminology, mitochondrial DNA, C-value
Introduction
General description
Amphibians (class Amphibia) are anamniotic tetrapods that include around 8,900 extant species worldwide. Most species spend their lifetimes split between land and water, hence the name amphibian, or ἀμφίβιος (amphíbios) in Greek.
A diverse array of fossil amphibians have been discovered, which are considered to be “primitive” tetrapods, most likely archaic amphibians, from the subclasses Temnospondyli and Lepospondyli and the family Albanerpetontidae (Matsumoto and Evans 2018). These animals include fossils with wide, stocky heads, and sharp teeth, some of them resembling crocodiles, lizards, and snakes. Ross (2014) classified 268 genera of fossil amphibians from 69 families and eight orders into 54 kinds. Many of these kinds are at the level of the family or the superfamily. Intriguingly, many families are monogeneric, because they contain only one species.
The term amphibian is sometimes mistakenly used to cover all “early tetrapods,” but this is incorrect. Rather, what are known today as amphibians include the subclass Lissamphibia, which are very different from fossil tetrapods in their morphology, ecology, and ethology. This group includes three groups: Anura/Salientia (frogs and toads, 7,838 species), Caudata/Urodela (salamanders and newts, 825 species), and Gymnophiona/Apoda (caecilians, 225 species). A possible fourth group of lissamphibians may be mentioned, namely Allocaudata, an extinct clade that superficially resembles salamanders.
Together, Anura and Caudata are known as the clade Batrachia. Most neobatrachians contain the same order of 39 genes on their mitochondrial genome (Zhang et al. 2013). The extant gene order for non-Batrachians (Gymnophiona) is the same as that of Batrachia, except that the tRNA for leucine, threonine, and proline are next to the tRNA for phenylalanine (Zhang et al. 2013). This is an indication that Gymnophiona is an apobaramin separate from Batrachia.
The word λισσός (lissós, or smooth) refers to the soft skin of lissamphibians that they use for cutaneous respiration (Kardong 2015, 106–108). Their skin contains a number of glands, including epidermal Leydig cells that exclude bacteria and viruses, and mucous glands, poison glands, and chromatophores in the dermis (Kardong 2015, 219–221).
No fossils have been found that definitively connect living amphibians with non-amphibians. The earliest fossil frogs show all the characteristics of modern frogs, including their saltatory system of locomotion (Kardong 2015 107).
Several traits separate frogs, salamanders, and caecilians from one another. These three groups could be classified into separate cognita on their different body plans (Sanders and Wise 2003).
Frogs generally reproduce by external fertilization and are characterized by a tadpole stage, which hatches from the egg and undergoes metamorphosis. They also have a special form of saltatory locomotion using large hind legs. Their skin can be bright and secret poisonous material. An eardrum, or tympanum is used to pick up vocalization cues. Toads differ from frogs based on their warty skin and raised glandular masses called parotoid glands behind their eyes. They are called Anura (no tail) because of their tailless, stocky body (Campbell and Reece 2005, 685–86; Kardong 2015, 107–108).
Salamanders are tailed amphibians, hence the name Caudata (tailed ones), with paired limbs. Many salamanders are more terrestrial, whereas newts split their time between land and water. Due to their terrestrial lifestyle, salamanders walk with a side-to-side motion. In contrast with frogs, salamanders lack eardrums. Reproduction can either be external or can happen via a spermatophore, a package of sperm placed on the ground by the male and then taken up by the female into her cloaca (Campbell and Reece 2005, 685–686; Kardong 2015, 107). Some species, such as brook salamanders have internal fertilization. Various species of Caudata, such as the axolotl (Ambystoma mexicanum), are of prime interest in cellular regeneration and the fight against aging (Bölük, Yavuz, and Demircan 2022; Huang et al. 2024).
Gymnophiona have several traits indicating genome decay since the Fall. They have slender, wormlike bodies and live exclusively in tropical environments. They lack feet, hence the secondary name Apoda (no feet). The skull is solid and compact, which suits their burrowing lifestyle. Fertilization happens internally, and viviparity is the most common way of breeding (Kardong 2015, 108). They have very reduced or absent eyes, with only a single photopigment with no color vision (Campbell and Reece 2005, 685–686; Wilkinson 2012). These animals also have a pair of special chemosensory organs called the tentacles, which are made up of parts of the eye and the animal’s vomeronasal organ (Pough 2007).
Previous analyses of amphibians
Based on the above considerations, Anura, Caudata, and Gymnophiona may be considered as three separate apobaramins. All three orders belong to their own cognita, according to Elder (2015). This study aims to break down these groups into further holobaramins.
In previous studies, Hennigan (2013a, 2013b) estimated 140 anuran, 53 caudate, and one gymnophionan kinds, all of which are extant. In addition, Ross (2014) enumerated 54 fossil amphibian kinds from 69 families representing 268 genera, raising the number of predicted amphibian kinds to 248.
Building upon these previous results, this study will focus on the analysis of genome size and mitochondrial DNA (mtDNA) sequence similarity. Despite the almost uniform body plan in these three groups, it is likely that many small amphibian kinds will be uncovered in the analysis, consistent with the morphological and fossil analysis.
Biblical Analysis
Genesis 1:20–25 describes the creation of marine animals, flying creatures and land animals on Days 5 and 6 of Creation Week. Since amphibians live on land and in the water, the biggest question concerning them is what day they were created on, Day 5 or 6? We must note that the term amphibian does not refer to a single kind, nor is it a concept found in the Hebrew Old Testament. Though they are tied to both land and water, in many groups one of the two lifestyles dominates.
Since there are multiple amphibian kinds, it is likely that some were created on Day 5, along with flying and swimming animals. Others, which are more terrestrial in their lifestyles, were likely created on Day 6.
Because the great majority of amphibians are at least partially tied to a terrestrial lifestyle, representatives of their kinds would have been on board the Ark during Noah’s Flood.
Materials and Methods
The orders Anura, Caudata, and Gymnophiona were analyzed separately, due to their disparate genome sizes and different morphologies. Since the level of the kind is around the level of the family, setting the order as the upper level of the kind instead of the class was justified.
All Supplementary data files and images are available on Zenodo at https://zenodo.org/records/16615864.
Data
The mitochondrial DNA (mtDNA) was downloaded for 362 amphibian species from the National Center for Biotechnology Information (NCBI) database. A list of species and the sequence similarity matrix can be found in Supplementary file 1. The mtDNA of six outlier species were also included. These include Latimeria menadoensis, Lepidosiren paradoxa, Neoceratodus forsteri, Protopterus aethiopicus, Protopterus annectens, and Protopterus dolloi. This raised the total number of species to 368.
Statistical tests
To discover well-defined holobaramins, it must be ensured that the similarity values calculated between all species pairs in the study follow the normal distribution as closely as possible. A normal distribution with a high average similarity value and a small standard deviation is the most optimal. In contrast, if two or more modes are present in the similarity distribution (multimodality), this may be an indication that two or more baramins are represented in the data.
The Hartigan dip test can be used to detect multiple modes in the data. In R, the “dip.test” method from the diptest package can be used on a vector of data. The value of the D-statistic tells us whether multimodality is likely or unlikely. A low D-value (paired with a high p-value) indicates unimodality. Similarly, the normally distributed similarity values must not be skewed. For this, the “skewness” method can tell us whether the normal distribution is skewed or not. The closer the skew is to 0, the less skewed the normal distribution is.
Software
The OGDRAW software (Greiner, Lehwark, and Bock 2019) was used to draw the mitochondrial genomes of Echinotriton andersoni (NC_017870.1) and Tylototriton verrucosus (NC_017871.1). The heatmaps and the sequence similarity histograms were drawn using R version 4.3.1. The heatmaps were drawn using the ward.D2 clustering algorithm. The number of species from all three groups was retrieved from AmphibiaWeb (AmphibiaWeb 2025). The mtDNA sequences were aligned using the MAFFT aligner (Katoh et al. 2002). Genome size differences were depicted using the Genome Size Analysis tool at The Molecular Baraminology Analysis Tool Suite (https://molbar.shinyapps.io/molbar).
Results
Since the three orders of amphibians are visibly distinct cognita, they were each analyzed separately. Genomically, Caudata has an extremely wide range of genome sizes, ranging from 10.12 pg (9.9 Gbp) to 120.6 pg (117.9 Gbp), if 1 pg corresponds to 978 Mbp (Gregory et al. 2006). These results are shown in table 1 and are visualized in fig. 1. Due to its genome size range and unique morphology, Caudata may be designated as an apobaramin. Likewise, due to their unique morphology, Anura and Gymnophiona can be designated as apobaramins. The genomes of anurans are also more highly methylated and contain more palindrome sequences (Beçak and Kobashi 2004).
When comparing C-value ranges between the three orders, the most significant difference was found between anurans and caudates, with a p-value of 3.7E-138. The difference between caudates and gymnophionans was also significant, with a p-value of 0.009. The difference in C-values between anurans and gymnophionans was not significant, with a p-value of 0.53. This means that these two groups need to be separated based on other criteria.
Anura
Based on the mitochondrial DNA results, 12 putative anuran holobaramins were predicted. The putative clusters and the corresponding statistics can be found in Supplementary file 2. The clusters are visible in fig. 2. The elbow plot showing the decrease in total within sum of squares can be seen in Supplementary fig. 1. The histogram showing the distributions of the sequence similarity values for all 12 candidate clusters can be found in Supplementary fig. 2. The Hopkins clustering statistic was 0.897, which indicates very good clustering.
Of these 12 groups, 11 had more than three members, and they were all statistically significant at the 5% level. Four groups had both a relatively low skewness (< 2.5) in their distribution of sequence similarity values. These correspond to the genera Bombina, Kaloula, Pelophylax, and Xenopus representing separate putative holobaramins. The genus Ptychadena had a low Dip value and a low skew.
The genus Bombina consists of the fire-bellied toads, which secrete poisonous chemicals on their brightly colored ventral regions. This genus is made up of two monobaramins, based on their body size (Pabijan et al. 2013). The large-bodied Bombina species are B. maxima and B. microdeladigitora, which live in Southeast Asia. The small-bodied species include B. bombina, B. fortinuptalis, B. lichuanensis, B. orientalis, and B. variegata, and live in Far East Asia and Europe.
Pelophylax forms a monophyletic group based on its mitochondrial genome, with three lineages, corresponding to three monobaramins. These are the P. kurtmuelleri, the P. shqipericus, and the P. nigromaculatus groups (Hofman et al. 2016).
Xenopus species all belong to their own holobaramin.
The genus Ptychadena consists of frog species widespread across Africa with a conserved morphology (Reyes-Velasco et al. 2018).
| Group | Minimum | Mean | Std. dev. | Maximum |
| Anura | 0.95 | 4.99 | 2.22 | 13.4 |
| Caudata | 10.12 | 35.8 | 17.2 | 120.6 |
| Gymnophiona | 3.7 | 7.45 | 5.65 | 13.95 |
Table 1. C-value statistics of the three extant amphibian groups.
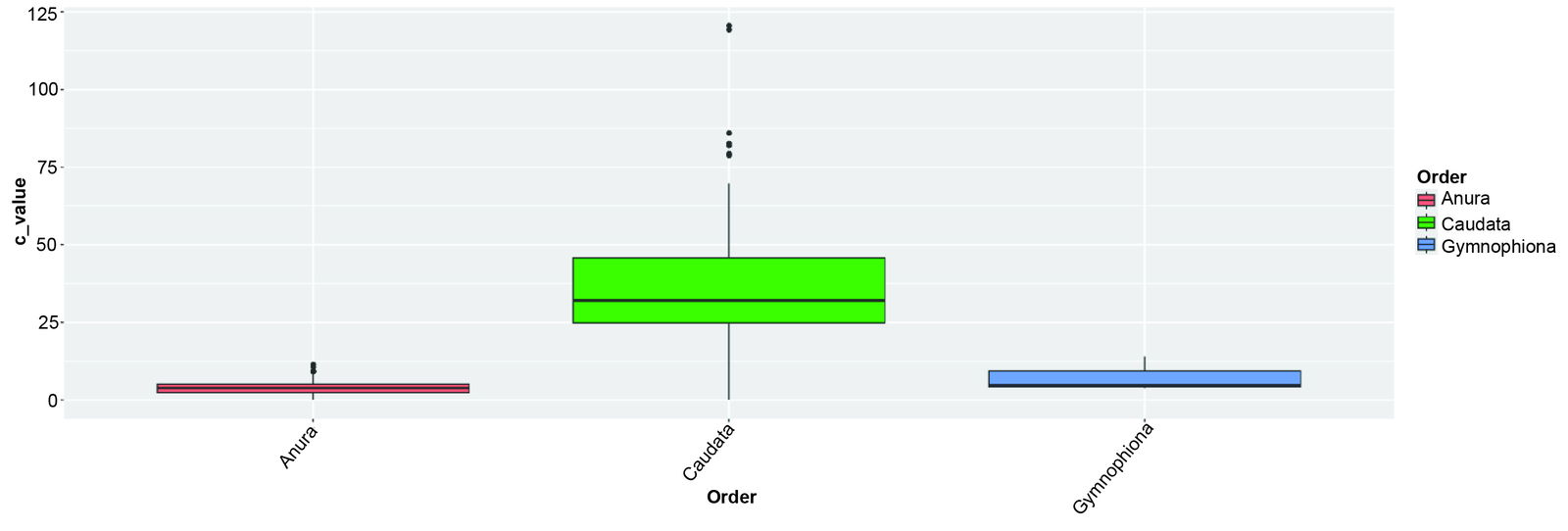
Fig. 1. C-value range of the three extant amphibian groups. Values taken from the Molecular Baraminology Analysis Tool Suite Database.
Caudata
The data appears to support seven putative Caudatan holobaramins. The putative clusters and the corresponding statistics can be found in Supplementary file 3. The clusters are visible in fig. 3. The elbow plot showing the decrease in total within sum of squares can be seen in Supplementary fig. 3. The histogram showing the distributions of the sequence similarity values for all 12 candidate clusters can be found in Supplementary fig. 4. The Hopkins clustering statistic is 0.865, which indicates good clustering.
However, only three groups had a low skew value (< 1.2) indicating unskewed, normal sequence similarity values. These three groups appear to be real holobaramins, corresponding to the genera Ambystoma (mole salamanders), Echinotriton + Tylototriton (the latter genus corresponds to crocodile or knobby newts), and Triturus (crested and marbled newts). These three groups were also found by Hennigan (2013b) in his study of amphibian ark kinds.
Eleven species from Ambystoma were analyzed. Interestingly, several species of Ambystoma are unisexual females, which sometimes steal sperm from sympatric sexual donor species. They reproduce via kleptogenesis, which means that they keep only some of the genetic material “stolen” in such a way from the sexual species (Bi and Bogart 2010). The unisexual species all contain a highly conserved mitochondrial genome, derived from A. barbouri (Robertson et al. 2006). Their ploidy levels can range from diploid to pentaploid. Because of the unusual mode of generation and inter-relation with sexual ambystomatids, it is likely that the genus Ambystoma is a holobaramin, with unisexual and sexual species both forming separate monobaramins.
The genera Echinotriton and Tylototriton consist of the crocodile and knobby newts. These two genera belong to the family Salamandridae, and the subfamily Pleurodelinae. The mean sequence similarity between E. andersoni (the Ryukyu spiny newt) and all Tylototriton species is 0.662 ± 0.002, whereas between all Tylototriton species the mean sequence similarity is 0.746 ± 0.041. What could be the reason for this difference? Upon closer inspection, there is a tandem 439 bp and a 324 bp repeat sequence in the T. verrucosus mitogenome (17,100 bp) compared to the mitogenome of E. andersoni, which is 16,268 bp. These repeats also contain an extra trnT (a CCA-adding enzyme) element between cytochrome-b and the control region (CR) (see red line in lower right of fig. 4). Furthermore, Kurabayashi et al. (2012) report that the trnT-trnP gene pairing is common among caudates, meaning that it can be used as an apobaraminic marker.
The genus Triturus consists of crested and marbled newts, which have a mostly terrestrial lifestyle. The crested newt species secrete powerful neurotoxins through their skin. Many Triturus species are parapatric (mutual geographic avoidance), although they readily hybridize with each other, which is a good indication of their holobaraminic status (Wielstra 2019). Interestingly, Triturus newts exhibit balanced lethal chromosomes, whereby chromosomes 1A and 1B balance each other out; heterozygous forms die in the embryo stage. This remains an evolutionary mystery as to how this genotype could have evolved, as it is present in various organisms from plants to insects (France et al. 2025).
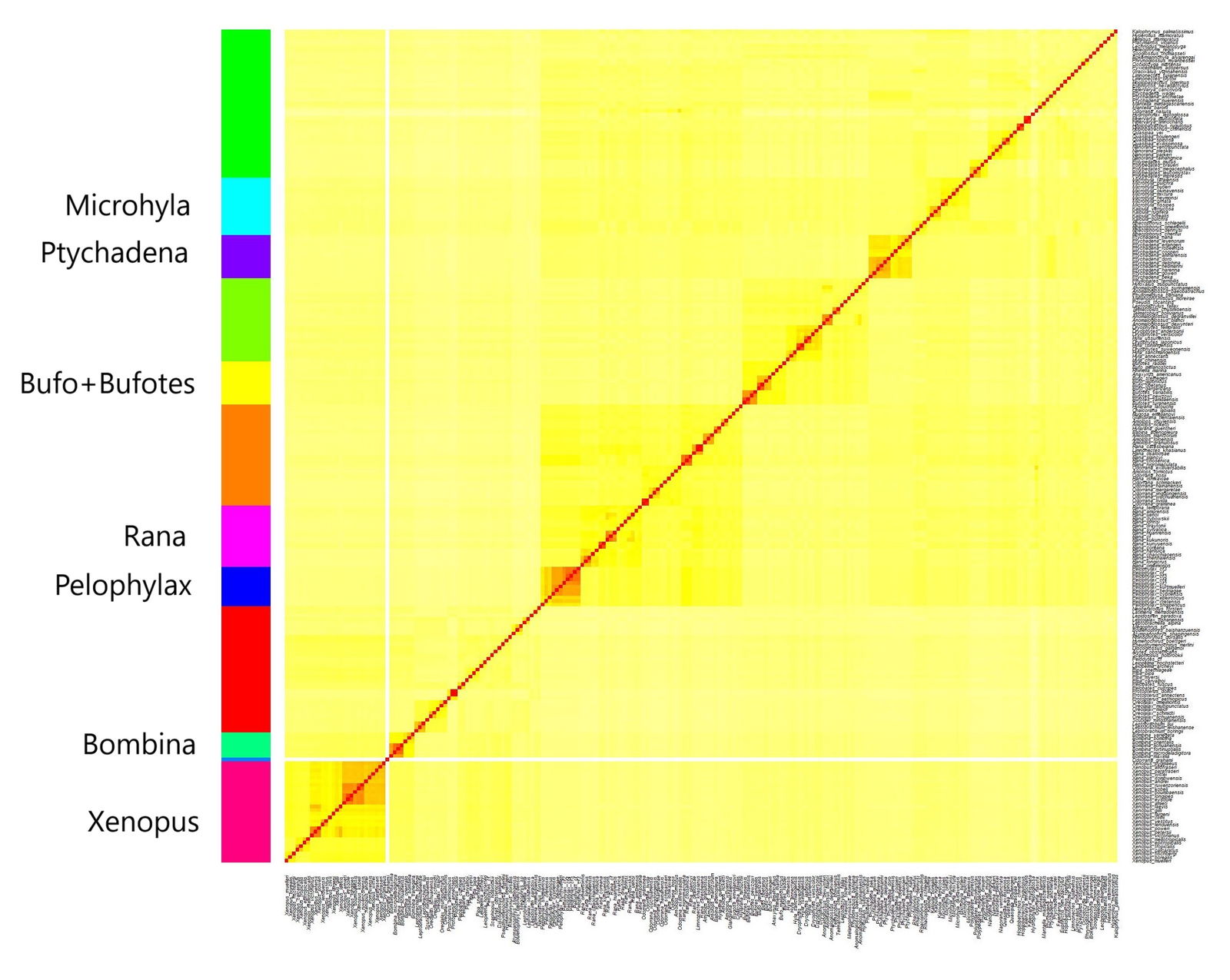
Fig. 2. Heatmap showing the 12 putative baramins discovered for Anura.
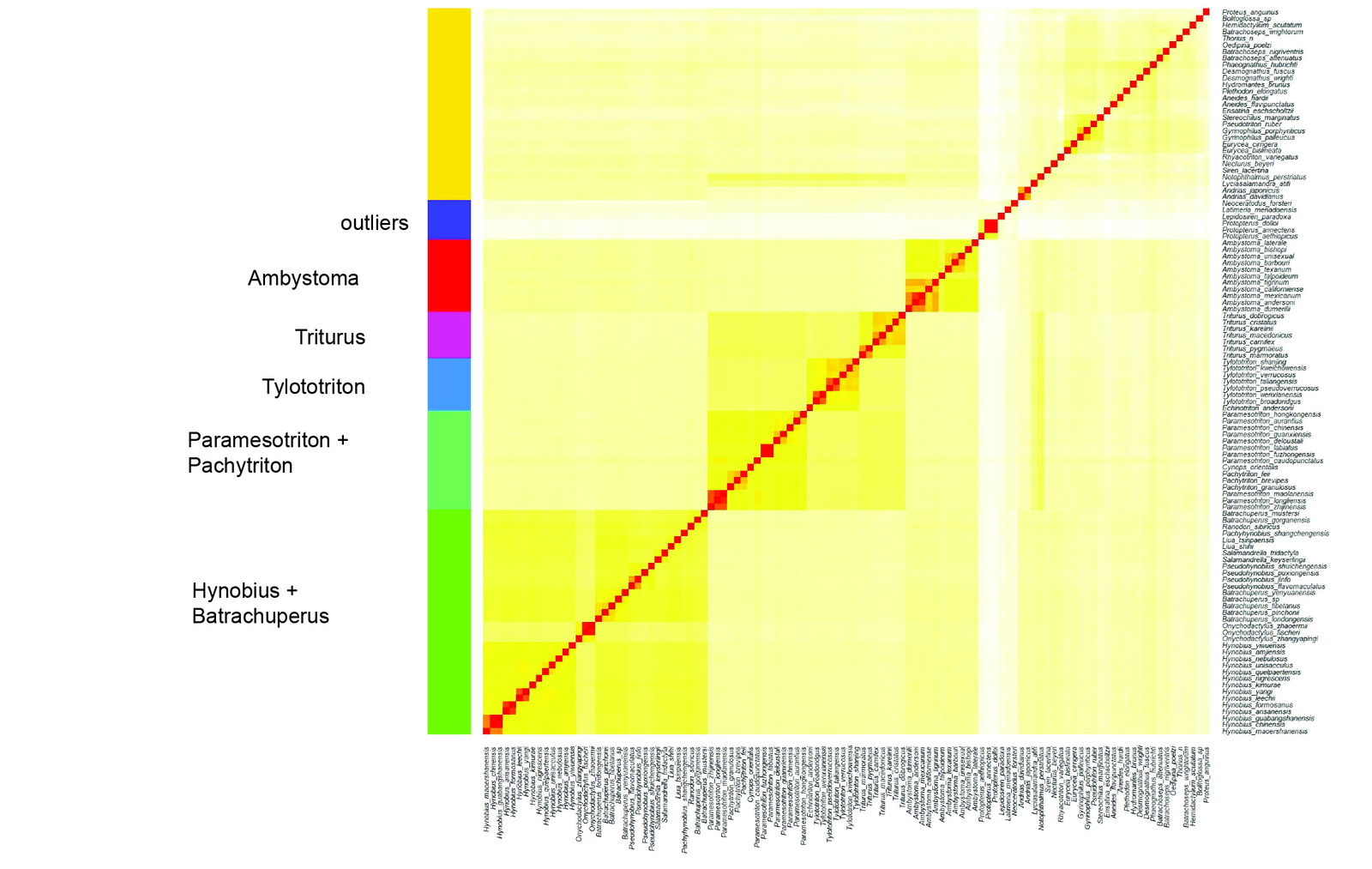
Fig. 3. Heatmap showing the seven putative baramins discovered for Caudata.
Gymnophiona
The results for Gymnophiona were somewhat puzzling. The putative clusters and the corresponding statistics can be found in Supplementary file 4. The clusters are visible in fig. 5. The elbow plot showing the decrease in total within sum of squares can be seen in Supplementary fig. 5. The histogram showing the distributions of the sequence similarity values for all twelve candidate clusters can be found in Supplementary fig. 6.
The Hopkins clustering statistic was 0.732, which indicates only fair clustering. The gymnophionans are clearly separated from the outlier groups Protopterus, Latimeria, Neoceratodus, and Lepidosiren. However, the elbow plot is a steadily decreasing curve without any inflection point (fig. 4a). It does not show any clear number of statistically significant groups. Putatively, 14 groups were analyzed, and all are statistically significant.
Three groups have a relatively low absolute skewness (< 1.24). These three groups are Caecilia + Oscaecilia (family Caeciliidae), Microcaecilia (family Siphonopidae), and three species from the genera Grandisonia, Hypogeophis, and Praslinia (family Grandisoniidae).
Despite these results, it may be possible that, in accordance with Hennigan (2013b), there is only one single gymnophionan holobaramin.
The biogeography of the ten families of Gymnophiona might reveal the baraminic status of these legless amphibians. Their largely sessile, burrowing lifestyle and lack of limbs would likely be very limited in crossing vast bodies of water between continents during uniformitarian geological conditions. But surprisingly, this is exactly what we find.
Table 2 shows the geographical distribution of the ten gymnophionan families. Gymnophionans are found in Central and South America, Africa, and Asia. Three families, namely Dermophiidae, Grandisoniidae, and Ichthyophiidae are found on two continents; species of Dermophiidae live on both sides of the Atlantic. Four families, Caeciliidae, Rhinatrematidae, Siphonopidae, and Typhlonectidae all live in northern parts of South America. It might be that these four families form a larger holobaramin due to their similar geographical localization. The wide geographical distribution of gymnophionans could also very well be due to one or many gymnophionan holobaramins passively dispersing in different directions in the post-Flood world.
Summary and Conclusion
Based on their physical traits, Anura, Caudata, and Gymnophiona can be separated into their own apobaramins. Caudata separates from the other two groups because of the discontinuity in genome size. Gymnophiona is separated from the other two groups because of its mitochondrial gene order.
Two new statistical measurements were introduced to help delineate statistically significant holobaramins. The Dip-test measures the modality of a given set of sequence similarity values. Multimodal data is an indication that there are at least two separate holobaramins in each cluster. Skewness also helps determine whether a set of sequence similarity values represents a single holobaramin.
Based on the present clustering analysis, four putative anuran holobaramins were predicted with close to zero skewness, and a low p-value. One anuran group also had both a low Dip value and a low skewness. These are groups that are all at the genus level: Ptychadena, Bombina, Kaloula, Pelophylax, and Xenopus. In Caudata, three such putative holobaramins were predicted: Echinotriton + Tylototriton, Triturus, and Ambystoma. Gymnophiona is likely considered to be a single holobaramin based on its fair clustering and lack of a clear-cut number of putative holobaramins in its elbow plot.
The genus Bombina was also predicted by Hennigan (2013a). However Xenopus was separated from the genera Hymenochirus, Pipa, and Pseudhymenochirus. Hennigan (2013b) named Triturus and Ambystoma as separate holobaramins as well as both Echinotriton and Tylototriton, which were lumped together in the present analysis.
Acknowledgements
The author would like to acknowledge Dr. Marcus Ross from Liberty University for his helpful remarks in understanding how to analyze amphibians.
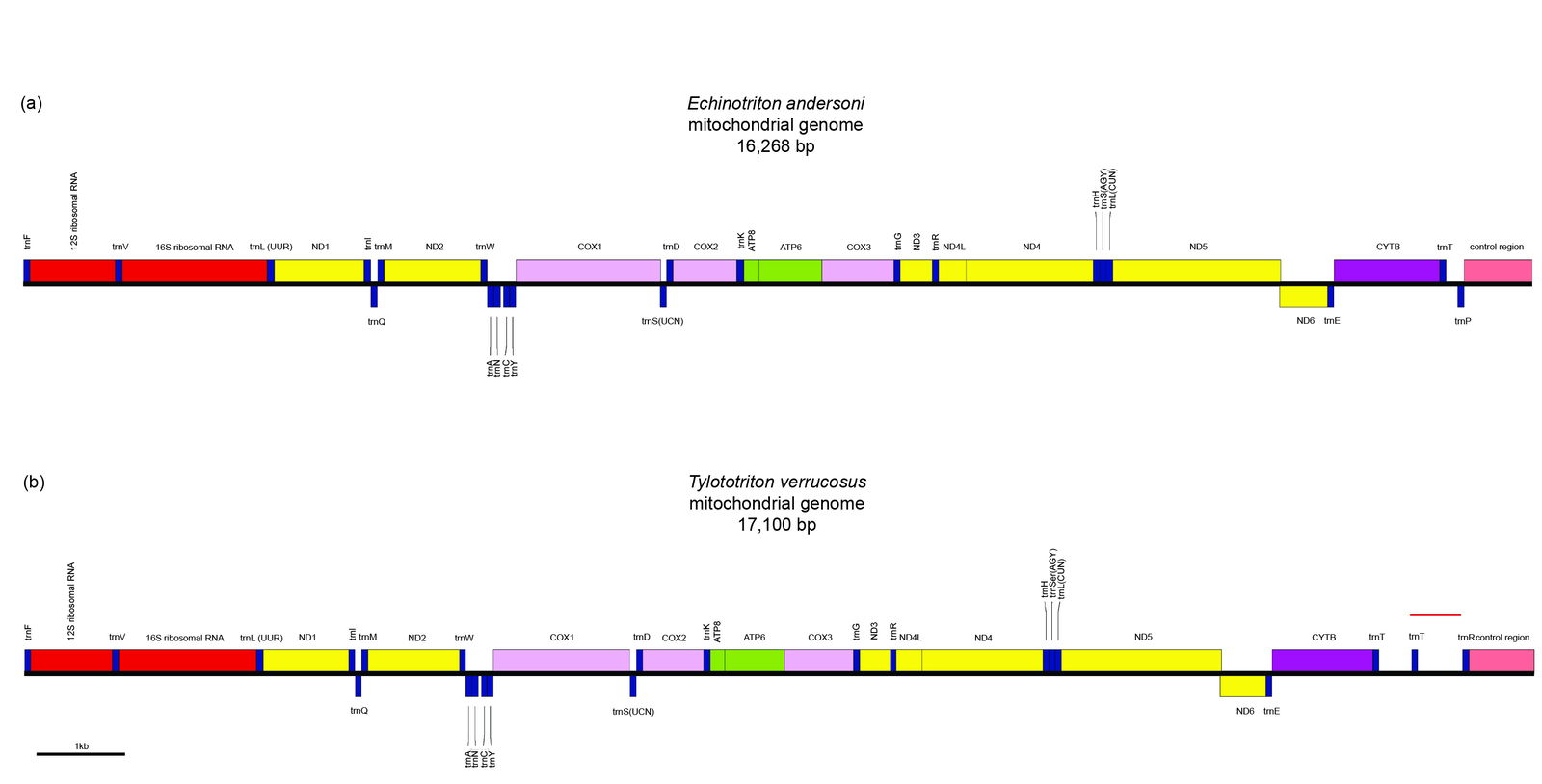
Fig. 4. Mitochondrial genome map showing the differences between the size and gene order of Echinotriton andersoni and Tylototriton verrucosus.
| Family | Number of species | Geographic distribution |
| Caeciliidae | 51 | Northern South America |
| Chikilidae | 4 | Small parts of northeast India |
| Dermophiidae | 15 | Africa, Central America, small parts of South America |
| Grandisoniidae | 24 | Seychelles, Cameroon, Ethiopia |
| Herpelidae | 11 | West Central Africa |
| Ichthyophiidae | 58 | Western coast of India, Indonesia, Southeast Asia |
| Rhinatrematidae | 14 | Parts of northern South America |
| Scolecomorphidae | 6 | Equatorial Africa |
| Siphonopidae | 28 | Northern South America |
| Typhlonectidae | 14 | Northern South America |
Table 2. Geographic distribution of families of Gymnophiona (data from the AmphibiaWeb Database).
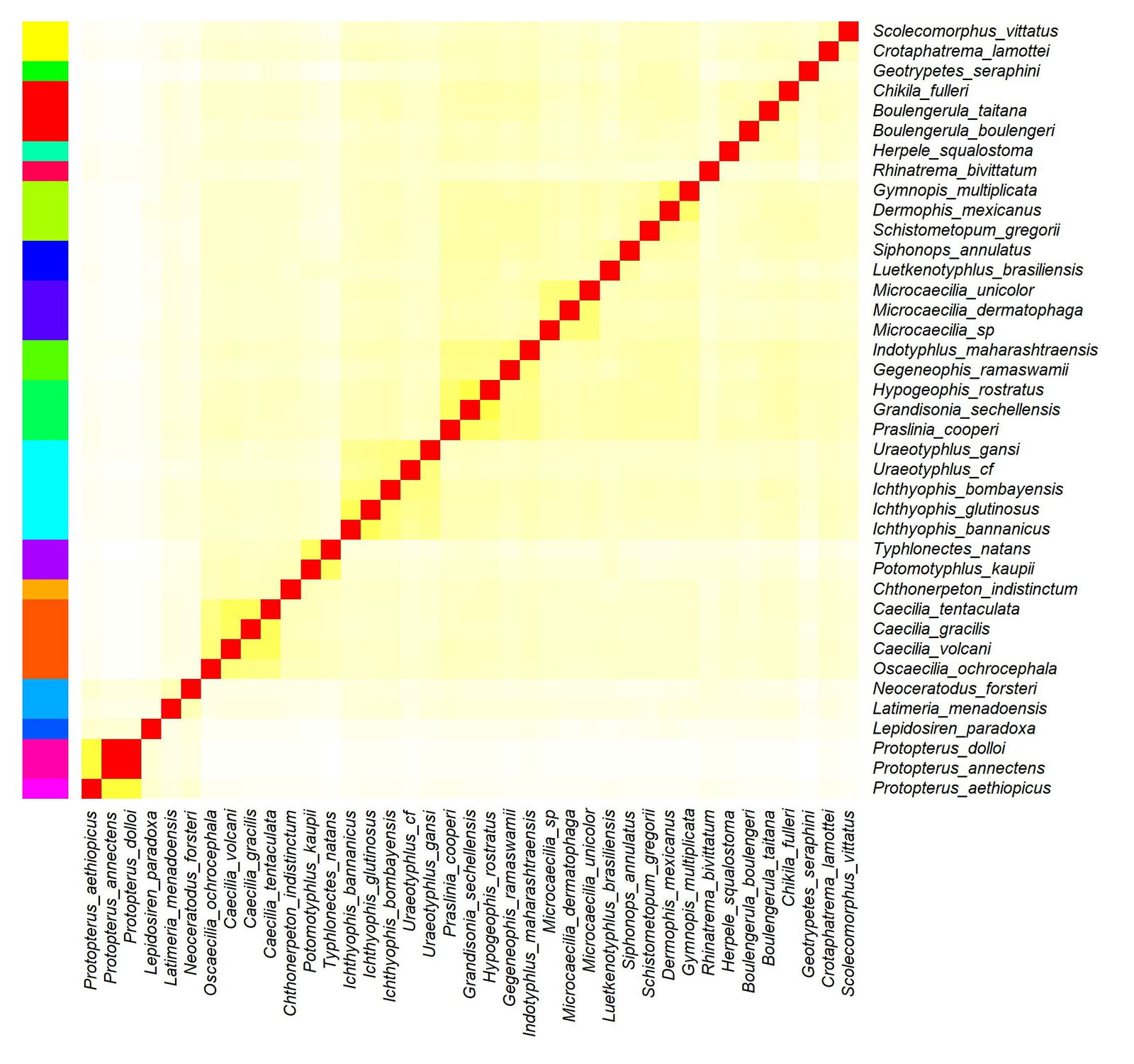
Fig. 5. Heatmap showing the 18 putative baramins discovered for Gymnophiona.
References
AmphibiaWeb. 2025. University of California: Berkeley, California. https://amphibiaweb.org.
Beçak, Maria Luiza, and Leonardo Setsuo Kobashi. 2004. “Evolution by Polyploidy and Gene Regulation in Anura.” Genetics and Molecular Research 3, no. 2 (June 30): 195–212.
Bi, Ke, and James P. Bogart. 2010. “Time and Time Again: Unisexual Salamanders (Genus Ambystoma) are the Oldest Unisexual Vertebrates.” BMC Evolutionary Biology 10, no. 238 (3 August): Article no. 238. https://doi.org/10.1186/1471-2148-10-238.
Bölük, Aydin, Mervenur Yavuz, and Turan Demircan. 2022. “Axolotl: A Resourceful Vertebrate Model for Regeneration and Beyond.” Developmental Dynamics 251, no. 12 (December): 1914–1933.
Campbell, Neil A., and Jane B. Reece. 2005. Biology. Pearson Education: San Francisco, California.
Elder, Todd W. 2015. Created Kinds, Baraminology and the Creation Orchard. Scripture Advocate Publishing: Livingston, Texas.
France, James, Manon Chantal de Visser, Jan W. Arntzen, Wiesław Babik, Milena Cvijanović, Ana Ivanović, Jeramiah Smith, Tijana Vučić, and Ben Wielstra. 2025. “The Balanced Lethal System in Triturus Newts Originated in an Instantaneous Speciation Event.” bioRxiv. https://doi.org/10.1101/2024.10.29.620207.
Gregory T. Ryan, James A. Nicol, Heidi Tamm, Bellis Kullman, Kaur Kullman, Ilia J. Leitch, Brian G. Murray, Donald F. Kapraun, Johann Greilhuber, and Michael D. Bennett. 2006. “Eukaryotic Genome Size Databases.” Nucleic Acids Research 35 (Database issue): D332-8. doi: 10.1093/nar/gkl828.
Greiner, Stephan, Pascal Lehwark, and Ralph Bock. 2019. “OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes.” Nucleic Acids Research 47, no. W1 (2 July): W59–W64.
Hennigan, Tom. 2013a. “An Initial Estimate Toward Identifying and Numbering the Frog Kinds on the Ark: Order Anura.” Answers Research Journal 6 (October 2): 335–365. https://answersresearchjournal.org/frog-kinds-order-anura/.
Hennigan, Tom. 2013b. “An Initial Estimate Toward Identifying and Numbering Amphibian Kinds Within the Orders Caudata and Gymnophiona.” Answers Research Journal 6 (January 23): 17–34. https://answersresearchjournal.org/amphibian-kinds-caudata-gymnophiona/.
Hofman, Sebastian, Maciej Pabijan, Artur Osikowski, Spartak N. Litvinchuk, and Jacek M. Szymura. 2016. “Phylogenetic Relationships Among Four New Complete Mitogenome Sequences of Pelophylax (Amphibia: Anura) From the Balkans and Cyprus.” Mitochondrial DNA. Part A, DNA Mapping, Sequencing, and Analysis 27, no. 5: 3434–3437.
Huang, Lu, Chiakang Ho, Xinran Ye, Ya Gao, Weiming Guo, Julie Chen, Jiaming Sun, Dongsheng Wen, Yangdan Liu, Yuxin Liu, Yifan Zhang, and Qinfeng Li. 2024. “Mechanisms and Translational Applications of Regeneration in Limbs: From Renewable Animals to Humans.” Annals of Anatomy 255 (August): Article no. 152288.
Kardong, Kenneth V. 2015. Vertebrates: Comparative Anatomy, Function, Evolution. McGraw-Hill Education: New York, New York.
Katoh, Kazutaka, Kazuharu Misawa, Kei-ichi Kuma, and Takashi Miyata. 2002. “MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform.” Nucleic Acids Research 30, no. 14 (July 15): 3059–3066.
Kurabayashi, Atsushi, Takuma Nishitani, Seiki Katsuren, Shohei Oumi, Masayuki Sumida. 2012. “Mitochondrial Genomes and Divergence Times of Crocodile Newts: Inter-Islands Distribution of Echinotriton andersoni and the Origin of a Unique Repetitive Sequence Found in Tylototriton mt Genomes.” Genes & Genetic Systems 87, no. 1: 39–51. https://doi.org/10.1266/ggs.87.39
Matsumoto Ryoko, and Susan E Evans. 2018. “The First Record of Albanerpetontid Amphibians (Amphibia: Albanerpetontidae) from East Asia.” PLoS One 13, no. 1 (January 3): e0189767.
Pabijan, Maciej, Anna Wandycz, Sebastian Hofman, Karolina Węcek, Marcin Piwczyński, and Jacek M. Szymura. 2013. “Complete Mitochondrial Genomes Resolve Phylogenetic Relationships Within Bombina (Anura: Bombinatoridae).” Molecular Phylogenetics and Evolution 69, no. 1 (October): 63–74.
Pough, Frank Harvey. 2007. “Amphibian Biology and Husbandry.” ILAR Journal 48, no. 3 (1 July): 203–213.
Reyes-Velasco, Jacobo, Joseph D. Manthey, Yann Bourgeois, Xenia Freilich, and Stéphane Boissinot. 2018. “Revisiting the Phylogeography, Demography and Taxonomy of the Frog Genus Ptychadena in the Ethiopian Highlands with the Use of Genome-Wide SNP data.” PLoS ONE 13, no. 2 (1 February): e0190440.
Robertson, Alexander V., Cadhla Ramsden, John Niedzwiecki, Jinzhong Fu, and James P. Bogart. 2006. “An Unexpected Recent Ancestor of Unisexual Ambystoma.” Molecular Ecology 15, no. 11 (October): 3339–3351.
Ross, Marcus. 2014. “Fossil Baramins on Noah’s Ark: The ‘Amphibians’.” Answers Research Journal 7 (September 17): 331–355. https://assets.answersresearchjournal.org/doc/v7/fossil-baramins-noahs-ark-amphibians.pdf.
Sanders, Roger W., and Kurt P. Wise. 2003. “The Cognitum: A Perception-Dependent Concept Needed in Baraminology.” In Proceedings of the Fifth International Conference on Creationism. Edited by Robert L. Ivey, 445–455. Pittsburgh, Pennsylvania: Creation Science Fellowship.
The Molecular Baraminology Analysis Tool Suite. 2025. https://molbar.shinyapps.io/molbar.
Wielstra, Ben. 2019. “Triturus Newts.” Current Biology 29, no. 4 (February 18): R110–R111. doi.org/10.1016/j.cub.2018.12.049
Wilkinson, Mark. 2012. “Caecilians.” Current Biology 22, no. 17: R668–R669. doi.org/10.1016/j.cub.2012.06.019
Zhang, Peng, Dan Liang, Rong-Li Mao, David M. Hillis, David B. Wake, and David C. Cannatella. 2013. “Efficient Sequencing of Anuran mtDNAs and a Mitogenomic Exploration of the Phylogeny and Evolution of Frogs.” Molecular Biology and Evolution 30, no. 8 (August): 1899–1915. https://doi.org/10.1093/molbev/mst091.







