The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
Staphylococcus aureus infections are a common cause of disease, particularly in colonized persons. Recently, a series of published articles have reported that community-acquired methicillin-resistant S. aureus (CA-MRSA) strains are evolving and increasingly becoming prevalent in households while health care acquired MRSA (HA-MRSA) is declining in the USA. The changing superbugs have often been used as an example of evolution in action. Although MRSA infections have become increasingly reported in the community, population-based studies of students preparing for the health professions having S. aureus and MRSA colonization are lacking. During the 2014–2015 school year 544 students were tested for MRSA carriage at Liberty University. A subset of the students, classified as having clinical exposure, had a 20%+ MRSA carriage rate which is 5–10 times the national average. We have seen a changing profile from HA-MRSA to CA-MRSA. This is potentially dangerous because the new strains are more virulent and aggressive. CA-MRSA is somewhat difficult to define—but these bacteria are mostly associated with their genetic and antibiotic profile (i.e. Cipro resistance), toxin genes, and place of acquisition. There are genetic variations of S. aureus strains among students of tightly knit groups: households, dorms, and other close living quarters. The bacteria are ping-ponging around among students, changing as they go. Since the majority of the students in the nursing and biology courses are looking to pursue a career in medicine, this sampling was beneficial to inform students, researchers, medical workers, and others if they are S. aureus and MRSA carriers, so they can take preventative measures to reduce the risk of infection.
Keywords: Staphylococcus aureus, MRSA, Staphylococcus carriage, clinical student risks, CA-MRSA, HA-MRSA, evolution of MRSA, superbugs
Changing Trends in MRSA
In the early years of our study, most nursing and advanced biology (BIOL 203 and 305) students carried mostly HA-MRSA due to their clinicals. However, this past school year students in the same classes primarily carried CA-MRSA isolates. Among biology majors taking general microbiology (BIOL 303), these students seldom carry MRSA and those who have it are CA-MRSA carriers. These students have a MRSA carriage rate like the general population in the USA (2%).
This change is real and clearly indicates an emergence of new MRSA variants that some may call microevolution. However, it is not Darwinian upward-onward evolution, but clearly adaptive changes within a species. They are variants on a theme. MRSA strains have longer doubling times (i.e. slower growth) than wild-type, less resistant, strains of S. aureus. These resistant bacteria have fitness costs to their changes: when more foreign genes are picked up via phage or plasmid acquisition, their growth rate diminishes. More research is needed to measure genetic change.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has captured national headlines. In the fall of 2007, the CDC (Center for Disease Control in Atlanta) reported that deaths due to MRSA were greater than those caused by AIDS for a number of years (Courvalin, LeClerq, and Rice 2011). MRSA is rapidly becoming one of the most prevalent and menacing diseases of our time and making time spent in a hospital an increasing risk. MRSA are bacteria that represent new strains of antibiotic resistant Staphylococcus aureus (figs. 1 and 2). Although the media discusses MRSA as a single new strain—in reality MRSA represents more than 1100 distinct strains of S. aureus (Wim and Neeling 2005). S. aureus causes many diseases (fig. 3), including skin boils, infection of leg ulcers, and pressure sores. Occasionally, it can cause more serious disorders such as blood infection (septicemia), pneumonia, bone, joint, or heart valve infection. The virulence factors and their resistance to antibiotics makes MRSA bacteria deadly. Methicillin is a penicillin derivative that was issued in clinics in most countries in 1959–1960; but discontinued after 1961 due to resistance strains appearing. In the late 1990s a resistance strain was found to be a community-acquired methicillin-resistant S. aureus strain. Unfortunately resistance has increased dramatically since 2000. Several national studies report that more people carry MRSA in communities than hospitals.
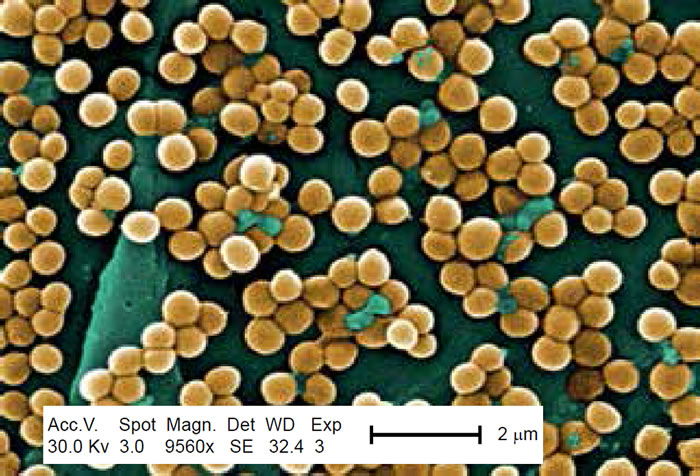
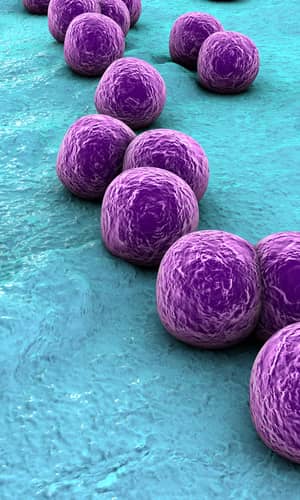
Figure 1. Scanning electron micrograph (SEM) depicting clumps of MRSA in gold, magnified 9560x. https://commons.wikimedia.org/wiki/Staphylococcus_aureus#/media/File:CDC-10046 MRSA.jpg (Gillen 2009).
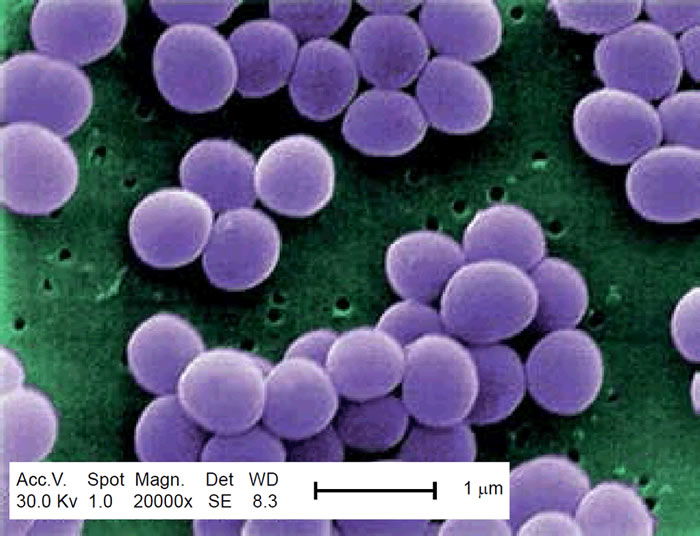
Figure 2. SEM depicting Staphylococcus aureus, in purple, magnified 20,000x. https://commons.wikimedia.org/wiki/File:Staphylococcus_aureus_VISA_2.jpg. (Gillen 2009).
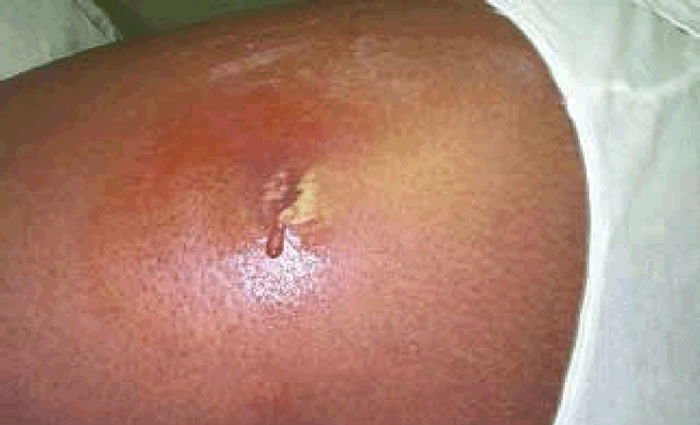
Figure 3. MRSA cutaneous abscess MRSA Staphylococcus aureus. http://commons.wikimedia.org/wiki/File:Cutaneous_abscess_MRSA_staphylococcus_aureus_7826_lores.jpg.
These bacteria are now categorized as hospital-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA) depending upon the disease acquisition location.
HA-MRSA and CA-MRSA are similar in many ways, but distinct. HA-MRSA resists many antibiotics but is sensitive to ciprofloxacin; whereas CA-MRSA resists few antibiotics including ciprofloxacin. Genetic traits for CA-MRSA include the Panton Valentine leukocidin (PVL), gene and a staphylococcal cassette chromosome of USA300 and USA400 types. CA-MRSA tests positive for hemolysins, leukocidin (PVL), and exfoliative toxin. HA-MRSA does not usually contain these toxins (table 1). It is this PVL toxin that makes the USA300 strain very dangerous. CA-MRSA commonly enters the blood stream through the lungs, surgical sites, and implant sites (table 2). However, it may also inflame a portion of the skin, appearing as a spider bite, pimple, or boil.
| MRSA in Health Care | MRSA in the Community | |
|---|---|---|
| Major Genotypes | USA100/200 | USA300/400 |
| Growth Rate | Slow compared to wild-type | Intermediate* |
| Antiobiotic Resistance | Many antibiotics | Few antibiotics |
| Cipro-sensitive | Cipro-resistant | |
| Virulence Factor Toxins | HA-MRSA | CA-MRSA |
| SCC Mec | Types I–III | Types IV, V |
| Cytolytic, ɑ | — | |
| Hemolysins | — | + |
| Leukocidin (PVL) | — | + |
| Exfoliative | — | some strains |
| Enzymes | ||
| β-lactamase | — | 90% of strains |
| Coagulase | + | + |
| Hyaluronidase | — | + |
| Staphylokinase | + | + |
| Phagocytic Inhibitors | ||
| Polysaccharide slime layer | + | + |
| CA-MRSA | HA-MRSA |
|---|---|
| Blood stream | Immunocompromised |
| Surgical site | Residency in long-term care facilities |
| Site of implant | Recent hospitalizations, recent surgery |
| Area affected skin and soft tissues | Dialysis patients, kidney, lungs causing pneumonia |
Although, HA-MRSA has various staphylococcal cassette chromosomes, USA100 and USA200 being most common. HA-MRSA commonly affects the immunocompromised, dialysis patients, postsurgical patients, people with recent hospitalizations, and patients in long-term care facilities (table 2). HA-MRSA is usually negative for cytolytic α hemolysins, PVL, and the exfoliative toxin. About 90% of CA-MRSA strains have β-lactamase and are positive for coagulase, hyaluronidase, staphylokinase, and cytolysins, whereas, HA-MRSA only contains the enzymes coagulase and staphylokinase (table 1). CA-MRSA is now becoming the dominant epivar in the USA. It is important to study from a health care point of view because most CA-MRSA isolates in the USA are more aggressive and faster growing than HA-MRSA.
The purpose of this article is to provide a reasonable explanation for the genesis, emergence, and dominance of community-associated MRSA. It also addresses the issue of whether this phenomenon is evolution in action. Microbiology research based on the creation paradigm appears to provide some answers to these puzzling questions regarding the new variants of Staphylococcus aureus and its new dominance in the United States.
Community-associated MRSA (CA-MRSA) was initially defined as infection of MRSA in an outpatient, or a patient released from the hospital or clinic in 24 hours, faster growing in vitro. However, it is now recognized that CA-MRSA has unique characteristics not related to hospitalization or time released from a health care facility. CA-MRSA has a unique genetic profile, antibiotic sensitivity/resistant pattern, epidemiology, presentation, and treatment (Weigelt 2008, 43). Ciprofloxacin (Cipro) resistance is due to the mutation of one gene base (Anderson 2005). We choose this trait as a marker because it is easy and relatively cheap to identify using the standard Kirby-Bauer disk diffusion test. In addition to Cipro, CA-MRSA is also resistant to clindamycin and erythromycin, yet is sensitive to most antibiotics in comparison to HA-MRSA.
A recent, major study (Alam et al. 2015) also reveals that a large proportion of the USA300 isolates sequenced are resistant to fluoroquinolone (ex. Cipro) antibiotics. The significance of their study is that if households serve as long-term reservoirs of USA300, household MRSA eradication programs may result in a uniquely effective control method.
Methods and Materials
The methods we used to do research are given more completely in a previous study (Gillen, Daycock, and Serafin 2014). Students were sampled at Liberty University in various microbiology classes to compare carriage of Staphylococcus sp. between those exposed to time in a clinical setting (one week or more) and those individuals not exposed to a clinical environment. In order to determine the prevalence of S. aureus carriers among individuals, several biology classes from Liberty University were examined. The bacterial isolates and culture swabs in this work were selected from those collected in our study of S. aureus and MRSA colonization in college students scheduled for microbiology class in their normal MRSA detection lab. Nasal swabs were obtained during lab time where students had already been instructed in sampling technique, plating, and growing bacteria (figs. 4–6). This sample collection protocol and survey study was approved by the Liberty University Institutional Review Board (IRB). It insured anonymity of all participants with an 11-question biomedical survey, results, and procedures. The primary means that determined the type of MRSA were through survey, interview, and antibiogram analyses (i.e. Cipro sensitivity).
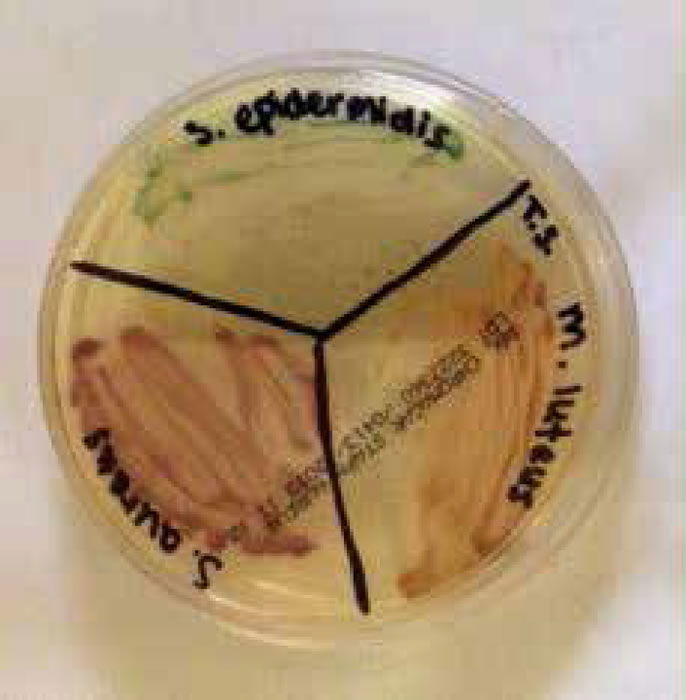
Figure 4. Staphylococci and micrococci colonies growing on CHROMagar™ Staph aureus (Gillen 2014).
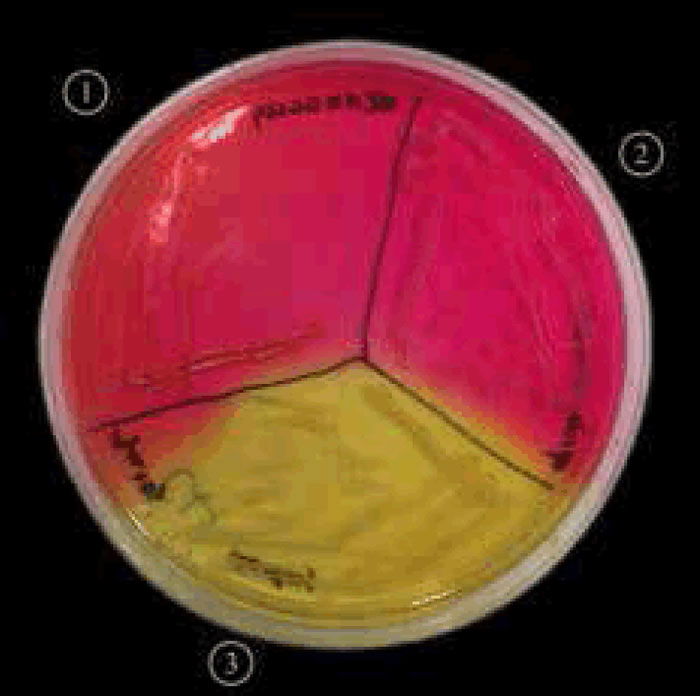
Figure 5. Staphylococcus aureus (MRSA) on mannitol salt agar with oxacillin.
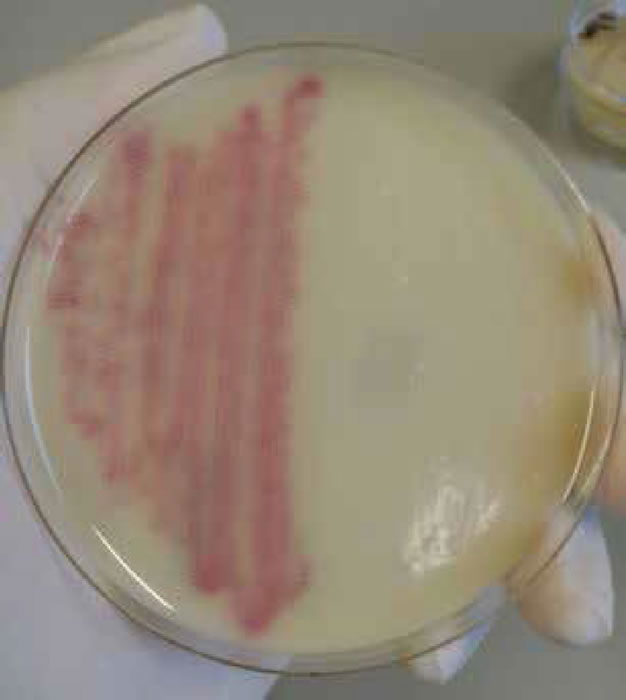
Figure 6. Staphylococcus aureus (MRSA) on MRSA select agar.
The student surveys were divided by academic class and section; the specific classes tested were Biology 203, 303, and 305 at Liberty University. The Biology 203 students consisted mainly of nursing and health promotion majors, in which the majority had exposure in a clinical setting. The Biology 303 and 305 students consisted mainly of biology majors, the majority of which have little direct exposure in clinical settings. The first step was to swab the student’s left and right nares as well as left and right axilla, using a separate, sterile cotton swab for each sample. The students streaked the contents from each swab onto a designated quadrant on a Petri plate with mannitol salt agar (MSA) which specifically tests for Staphylococcus growth. MSA contains a high concentration (7.5%) of salt, making it selective for members of the family Micrococcaceae and Staphylococcus, since this level of NaCl is inhibitory to most other bacteria. The plates were incubated at 37°C for 48 hours. In addition to obtaining a culture sample, each individual was asked to complete an anonymous survey with a correlated number to place on his or her plates. The survey included questions that could lead to potential reasons for carriage.
Once the incubation was complete, the plates were examined for any bacterial growth. If yellow cultures were present on the MSA, this marked a positive for S. aureus growth. If the MSA turned yellow it indicated mannitol fermentation, a common trait of S. aureus. The Staphylococcus culture was then transferred onto a MSA plate containing 4–6 μg/mL oxacillin to test for the presence of MRSA. Oxacillin, which is in the same class of drugs as methicillin, was chosen as the agent of choice for testing due to its chemical stability. For those MRSA positive, we also followed Kirby-Bauer procedure for antibiotic sensitivity using Mueller-Hinton plates.
CHROMagar™ Staph aureus, and MRSA Select were also agar types used for determining questionable MSA results and clarifying Staph and MRSA colony variations. Both agar types are chromogenic and display various color intensities that predict variation among S. aureus biotypes (or strains). We also tested for Coagulase Negative Staphylococcus (CoNS) bacteria in all sections as a control.
Results
In the BIOL 203 (clinically active nursing microbiology), 16% of students during the 2013–2014 school year were positive for MRSA (fig. 7), as opposed to 19% in the 2014–2015 school year. In the BIOL 303 (majors microbiology), 2% tested positive for MRSA during 2013 versus 17% in 2014. The reason for change was the 2014 class had high clinical exposure. In the BIOL 305 class (advanced microbiology and parasitology), 17% of students tested positive for MRSA during the 2013–2014 school year, and 30% in the 2014–2015 school year (fig. 8).
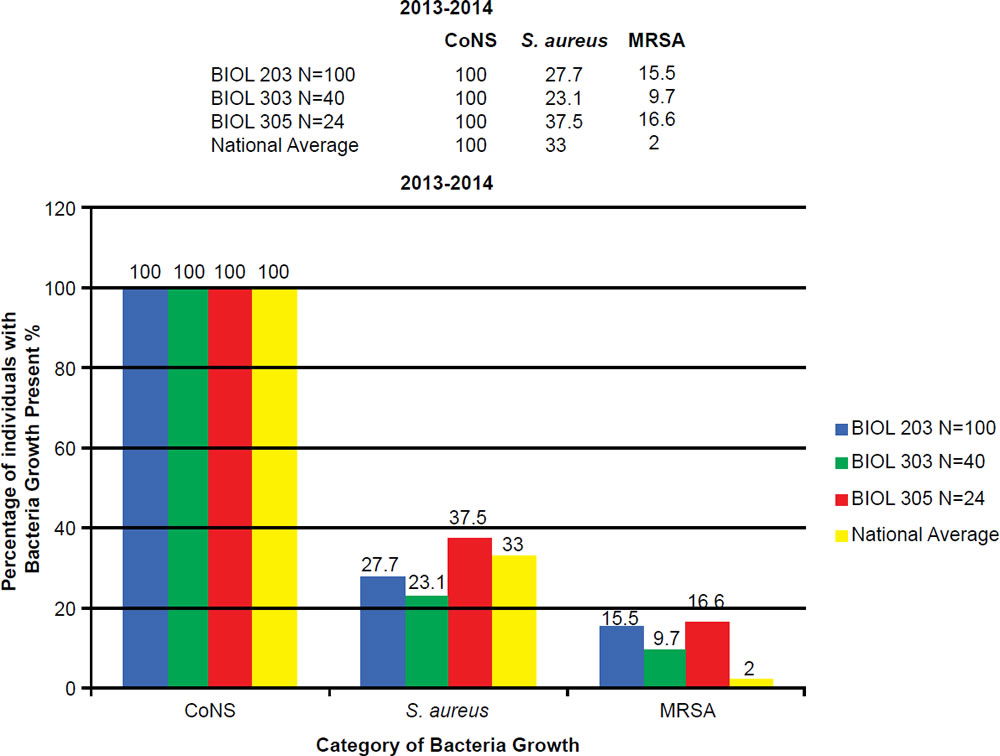
Figure 7. 2013–2014 Abundance of Coagulase Negative (CoNS), Staphylococcus aureus, and MRSA.
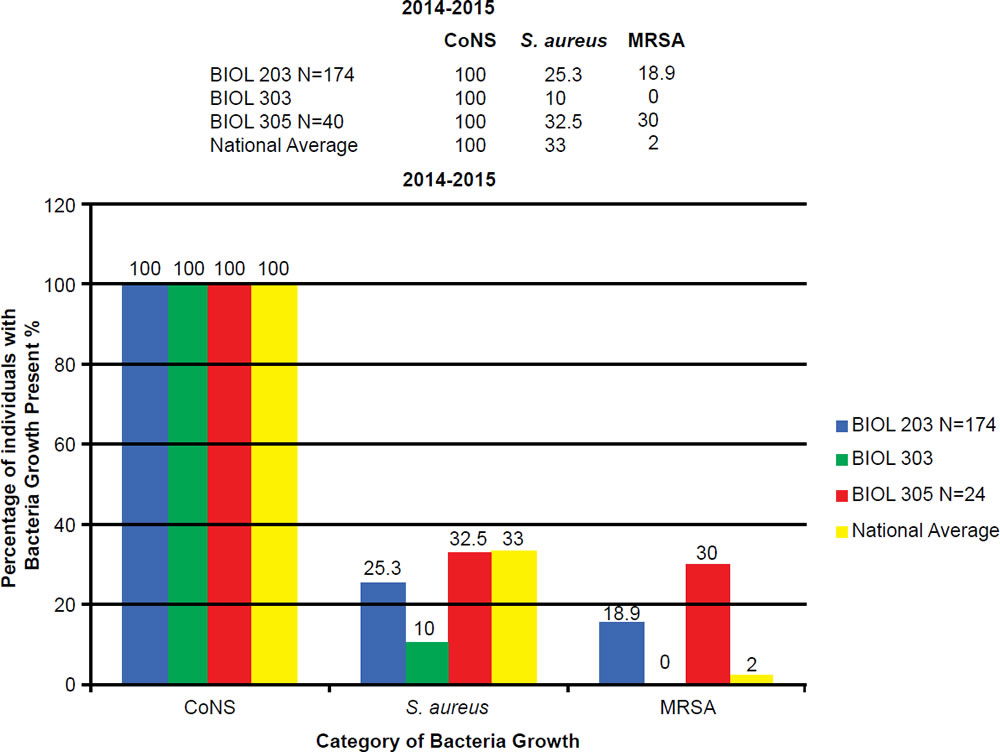
Figure 8. 2014–2015 Abundance of Coagulase Negative (CoNS), Staphylococcus aureus, and MRSA.
BIOL 203, BIOL 303, and BIOL 305 students have a 5 to 30 times higher MRSA carriage rate than the national average. The prevalence of MRSA among BIOL 203 and BIOL 305 students has been on the rise at Liberty University. This is largely due to high interest in the health professions, working at clinics, or being around people in the community that have MRSA.
At Liberty University, we sampled 544 students and had a high MRSA carriage rate common among clinically-oriented students, five to ten times the national average, which is 2% (figs. 7 and 8). In particular, we compared BIOL 203 students carrying MRSA in 2013–2014 versus 2014–2015 and found significantly more CA-MRSA this past year. In 2013–2014, we found 10 of 32 MRSA carriers (N = 206 sampled students) had CA-MRSA; whereas, in BIOL 2014–2015 we found 14 of 19 MRSA carriers (N = 174 sampled students) had CA-MRSA. There was a statistical difference in the type of MRSA carriage rate between the two sampling years (χ2, p = 0.0033). We have also seen a changing profile from HA-MRSA to CA-MRSA, which are potentially dangerous new strains more virulent and aggressive (table 3).
| Antibiotic | MRSA 10 | MRSA 15 | MRSA 2 | MRSA 18 | MRSA 27 | MRSA 4 | MRSA DA | F182 |
|---|---|---|---|---|---|---|---|---|
| CIP5 (5 μg) | 10 mm | 10 mm | 8 mm | 10 mm | 8 mm | 0 mm | 0 mm | 20 mm |
| Antibiotic | MRSA 14 | MRSA 3 | MRSA 26 | MRSA 12 | MRSA 13 | MRSA 16 | MRSA 7 | Average |
| CIP5 (5 μg) | 0 mm | 8 mm | 0 mm | 8 mm | 8 mm | 0 mm | 8 mm | 7.0 mm |
Alexandra Sheeran (2014) sampled 332 dorm students at Liberty University and found that students living in close-knit dorms (community shared bathrooms, etc.) had a much higher MRSA carriage rate (8.4%) than those in spaced dorms (1.5%). Those in spaced dorms had a rate similar to the national average (2%). While the large health care worker population may play a part in this trend, the rise in CA-MRSA carriage indicates a change in MRSA trend. We have also seen a changing profile from HA-MRSA to CA-MRSA, which are potentially dangerous new strains more virulent and aggressive.
Discussion
Liberty Research Verifies National Study
There was an overall trend change of MRSA among the Liberty University student population. A shift from HA-MRSA to CA-MRSA among the Liberty University student body over the 2013–2014 and 2014–2015 school years is evident after testing (figs. 7 and 8). There has been a rise in CA-MRSA from 2013 to 2015. The surveys indicated the prevalence of HA-MRSA among nursing majors with clinical experience, while other carriers who lacked clinical experience typically harbored CA-MRSA. Distinguishing HA-MRSA from CA-MRSA was accomplished by discovering the strain’s antibiotype through a Kirby-Bauer test. CA-MRSA was resistant to ciprofloxacin, whereas HA-MRSA was susceptible to ciprofloxacin (figs. 9 and 10).
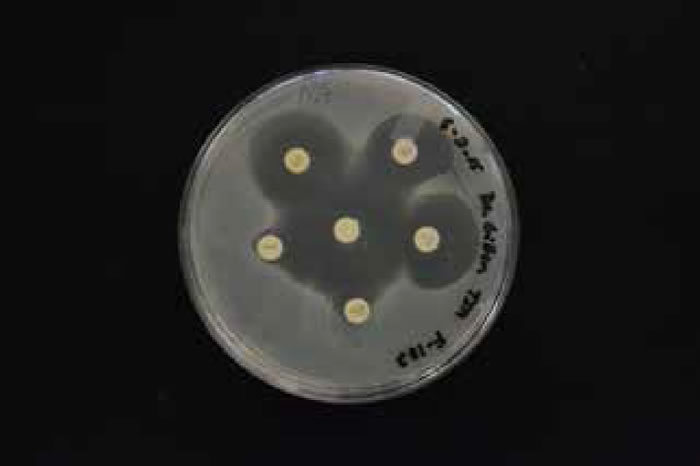
Figure 9. HA-MRSA photo antibiogram 1 of F-182.
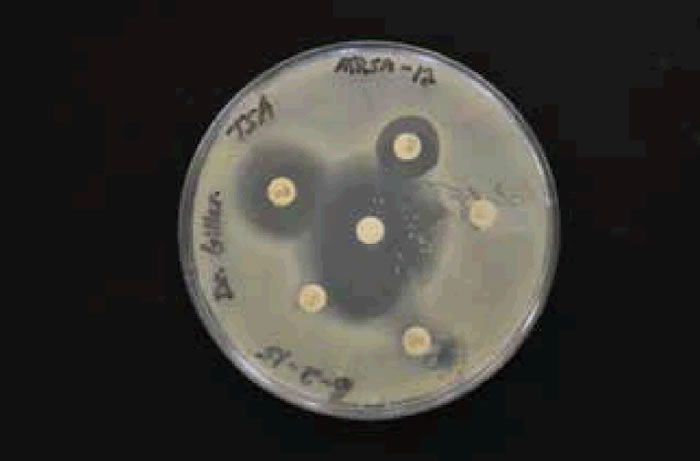
Figure 10. CA-MRSA photo or antibiogram 2 of student with CA-MRSA.
Our conclusion coincides with a study done by the University of Chicago that analyzed a growing trend of CA-MRSA (USA300) carriage in major cities across the United States of America (Alam et al. 2015). The rapid change within CA-MRSA could be explained by the ping-pong phenomenon (fig. 11). If the ball hits a paddle that has a sticky gene, then hits another with a second gene, and finally, a third gene, the end result would be all genes attached to the ball. In similar fashion, if a S. aureus carrying unique bacterial DNA enters one person, then two more unique variants invade a second and third person in the same family, the individuals of this family may carry any combination of the three new DNA strips, and thus, different MRSA isolates (fig. 11). In like manner, a similar pattern may be happening in college close-knit dorms, like those at Liberty University.
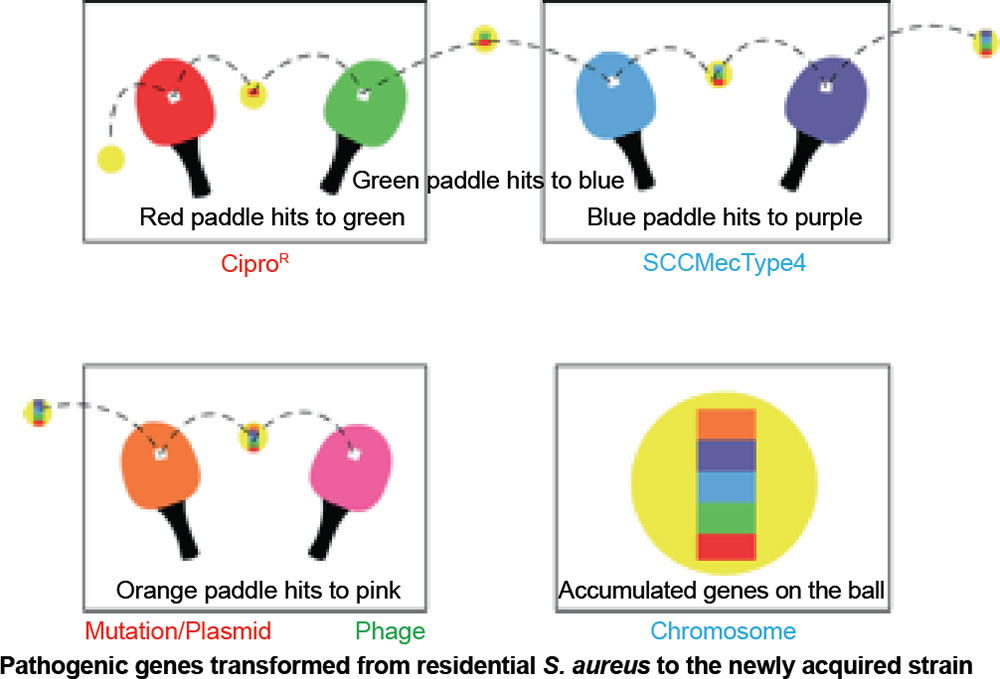
Figure 11. Ping-pong phenomenon: Ping-pong MRSA evolution. Table tennis (ping-pong) is an illustration of genetic change in MRSA species. If the ball hits a paddle with a sticky red strip, then hits another with a green strip, and finally, a blue strip, the end result would be a red, blue, and green strip all attached to the ball and so on. In similar fashion, if a S. aureus coccus enters one person carrying unique DNA, then a second, and third person, the final person in a family may carry all three DNA strips, and thus, different MRSA isolates. This research adds to the knowledge base of how USA300 MRSA spreads throughout the country, and hints at how it emerges inside households and other shared spaces (gyms, etc). In like manner, a similar pattern may be happening in college close-knit dorms, like those at Liberty University.
At Liberty University, there is a higher concentration of health care workers than the general American public. Despite this, the rising trend in CA-MRSA is still evident as Liberty University serves as a microcosm for the American public.
In the ASM mBio journal, Alam et al. (2015) reported that they evaluated the samples to understand transmission dynamics, genetic relatedness, and microevolution of USA300 MRSA within households. For the study, researchers used genome sequencing on 146 USA300 MRSA samples. These samples were collected during a previous study from 21 households in Chicago and Los Angeles where a family member had presented to the emergency room with a skin infection that was caused by USA300 MRSA. They also compared genetic information from these MRSA samples with previously published genome sequences of 35 USA300 MRSA isolates from San Diego and 277 USA300 MRSA isolates from New York City, as well as with the completed genomes of the bacteria USA300 variants. They created an evolutionary tree (fig. 12) to show the relationships among the bacterial strains. From a creation perspective, this tree shows variation and adaptation of S. aureus over the last decade.
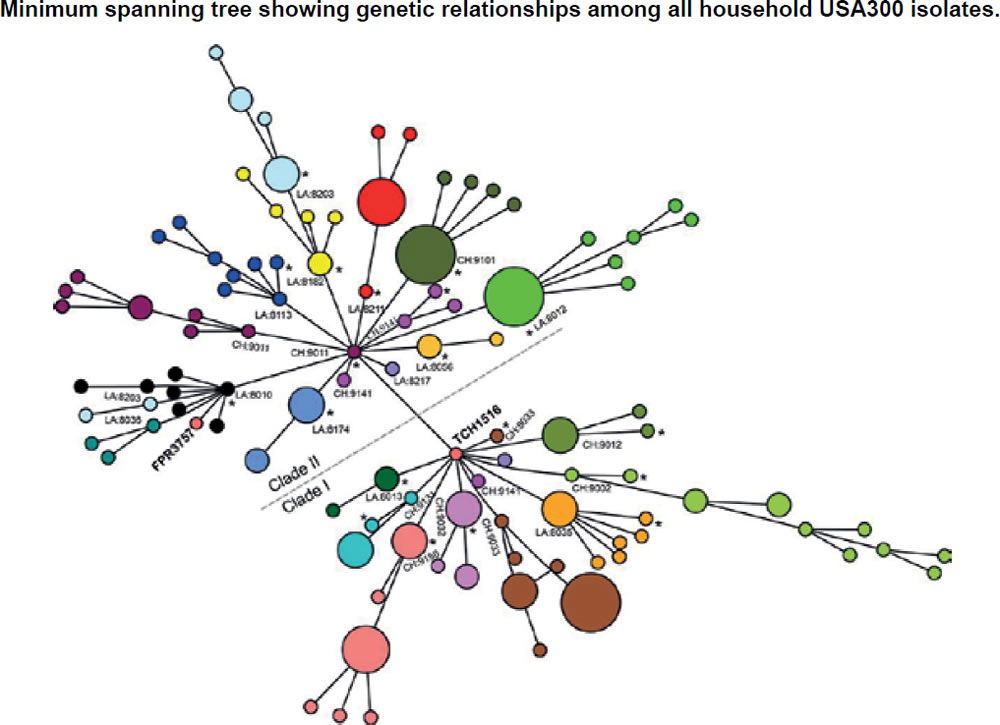
Figure 12. Evolutionary tree for CA-MRSA (after Alam et al. 2015).
These Chicago researchers found that isolates within households clustered into closely related groups, suggesting a single common USA300 ancestral strain was introduced to and transmitted within each household. Researchers also determined from a technique using evolutionary reconstruction that USA300 MRSA persisted within households from two to eight years before their samples were collected, and that in the course of a year, USA300 strains had a one in a million chance of having a random genetic change as a baseline, estimating the speed of change in these strains. Researchers also found evidence that USA300 clones, when persisting in households, continued to acquire extraneous DNA.
The NIH study (Alam et al. 2015) also reported that USA300 MRSA strains within households were more similar to each other than those from different households. Although MRSA is rarely introduced into households, it can persist there for years, ping-ponging around from person to person. The NIH findings strongly suggest that unique USA300 MRSA isolates are transmitted within households that contain an individual with a skin infection (fig. 12).
Multiple Isolates (Strains, Biotypes)
One person can harbor different variations of S. aureus. Upon examination of five people, we found two or more MRSA antibiograms based upon colony color, antibiotic sensitivity, and resistance. Two unique strains that were isolated from separate nares of the same person were sensitive to antibiotics differently. The antibiogram patterns were distinct indicating various biotypes or strains. Some examples are given in a previous paper (Gillen, Daycock, and Serafin 2014). The multiple isolates are likely due to numerous exposures to various people and the ping-pong phenomena occurring in dormitories, households, athletic facilities, and clinics.
Public knowledge of this shift is important as MRSA carriage can have serious health effects if the bacteria become invasive. The MRSA threat is trifold. First, carriers can pose a threat to individuals with compromised immune systems. Second, the carriers can endanger themselves by carrying MRSA should they fall sick and have a compromised immune system. Third, the carrier could aid the spread and change of the MRSA strain should they not seek treatment. Data show that S. aureus is changing, and a carrier could further the change by serving as a platform for the ping-pong phenomenon to occur. Early detection and treatment would prevent these negative outcomes from occurring.
Genetic Change
CA-MRSA contains a virulence factor that destroys leukocytes. The PVL toxin Panton Valentine gene facilitates destruction of neutrophils, macrophages, and in turn soft tissues and skin. A new bacteriophage (LH1) was isolated from raw milk using a Staphylococcus aureus ST352 host. Its genome includes the two genes encoding Panton-Valentine leukocidin (PVL), a virulence factor usually harbored by S. aureus prophages. A phage lytic mutant was isolated, and its analysis revealed the deletion of genes coding for the PVL and an integrase. The gene loss likely occurred through recombination between direct repeats (Haddad and Moineau 2013).
The genetic change is real and indicates the emergence of new MRSA variants, which some may describe as microevolution. However, this change is not upward, onward Darwinian evolution, but rather, an adaptive change as the species adjusts to its environment. Random changes that occur as patients harboring MRSA strains are exposed to other MRSA isolates that can then share genetic traits. This reshaping could be due to mutation, but the rapid diversification of MRSA among households (through the ping-pong effect) indicates that horizontal transfer through plasmid, or further yet, phage, is occurring. Again, it must be understood, while the change makes new strains of MRSA more virulent and aggressive, this is not a case of molecules-to-man evolution. MRSA is weaker in a sense than its wildtype (less resistant) counterpart as it has a longer doubling time than non-resistant S. aureus (Ender et al. 2004). In order to survive against antibiotics, strains of MRSA that resist antibiotics have been selected at the cost of their own fitness. As MRSA picks up more genes to survive against antibiotics, it sacrifices some of its fitness (more foreign genes acquired results in slower growth) to survive in its environment. The new genes are a temporary beneficial change for the bacteria, an adaptation in order to survive in a hostile environment.
Creation and MRSA
Where do the CA-MRSA microbes fit into God’s very good creation? Most creation biologists believe that God created (Genesis 1 and 2) all bacteria as very good forms of life (Gillen 2009, 2015). The Creator most likely wove1 together man’s skin with a Staphylococcus kind. Perhaps, the original Staphylococcus kind diversified into several varieties according to microhabitat: some to live on the dry skin (primarily Staphylococcus epidermis) and some to live on moist skin, like Staphylococcus aureus, which frequently is carried harmlessly in the nose and occasionally colonizes the skin without harm (Gillen, Daycock, and Serafin 2014).
Since the Fall, many bacteriophages and other genetic elements (plasmids) have invaded the original very good bacteria that are actually beneficial to us. Those toxins that are useful in the environment, having been displaced to the wrong location (that is the human body), have become dangerous. Super S. aureus evades the body defenses by secreting various enzymes to gain further expansion into the human body. For CA-MRSA, their toxins include leukocidins. The most dominant strains secrete a Panton Valentine leukocidin (PVL). PVL is associated with the majority of CA-MRSA strains in the USA and in particular MRSA USA300 strains. In addition, the added DNA codes for toxins and injurious chemicals that facilitate its spread across the skin, mucous membranes, and other body surfaces. The consequences to the Staph invasion can then lead to harmful effects as discussed already (Tortora, Funke, and Case 2015).
CA-MRSA started about 1997. Community-associated MRSA infections are transmitted within a community that are not health care related. But, they can enter hospitals and are doing so. In the Chicago research study (Alam et al. 2015), they mostly looked at households. But, the same seems to be occurring in athletic facilities and close-knit dorms. Anytime there is skin contact from a carrier, or a fomite from a carrier, it can transmit. Extensive contact with lotions, soaps, cosmetics, towels, razors, hairbrushes, nail files, and athletic equipment increases risk of contraction. Those athletes in close physical contact (contact sports) have a higher risk. Those that have open cuts, wounds, etc. are even more susceptible. Those that visit clinics or hospitals have an even at higher risk both to themselves and to the community. At Liberty University, a number of student-athletes are in clinical majors such as nursing, health promotions, and biomedical majors. This may be one of the reasons for high MRSA carriage among students in our microbiology classes.
In CA-MRSA, lateral transfer of DNA may be beneficial to the bacteria for a season. Virulence seems to commonly arise via mechanisms of conjugation, pathogenicity island transfer, bacteriophage insertion, and loss of metabolic coding information (fig. 11). For example, bacteriophages (viruses that infect S. aureus) are known to carry incomplete or corrupted genes that are then spliced into the genome of the host they infect. In addition, pathogenicity islands via conjugation have been transferred from other bacteria adding new virulence factors (fig. 11). The most significant lateral transfer appears to be the addition of chromosomal DNA cassettes (pathogenicity islands) from neighboring bacteria (Seifert and DiRita 2006).
But, super S. aureus evades the body defenses by secreting various enzymes to gain further expansion into the human body. Since the Fall, many bacteriophages and other plasmids have invaded Staph. For CA-MRSA, their toxins include hemolysins and leukocidins. The most dominant ones are those that secrete a Panton Valentine leukocidin (PVL) that destroys leukocytes. PVL is associated with the majority of CA-MRSA strains in the USA and MRSA USA300 strains in particular (figs. 11 and 12). In addition, the added DNA codes for toxins and injurious chemicals that facilitate its spread across the skin, mucous membranes, and other body surfaces. The consequences to the Staph invasion can lead to harmful pneumonia, impetigo, flesh-eating effects, and septicemia (Tortora, Funke, and Case 2015).
Evolution in Action?
According to most secular microbiologists, the type IV SCCmec may be one of the fit SCCmec types that can confer β-lactam resistance to community strains of S. aureus without greatly compromising their competitiveness among the natural flora of humans. It helps with fitness of CA-MRSA; i.e. less baggage of genes weight enables faster growth times than HA-MRSA; but still slower than original S. aureus wild-type (Ender et al. 2004). Darwinian evolutionists attempt to offer explanations on antibiotic resistance and prescriptions for future drug development. If they simply suggest an awareness of ongoing changes in pathogenic bacteria, we would concur. Bacteria do acquire resistance quickly, and many older drugs no longer work in hospitals and clinics.
The results of our previous experiments do show that bacteria can change quickly (Gillen and Anderson 2008). Although the acquisition of antibiotic resistance does not demonstrate Darwinian evolution, it does demonstrate that bacteria were endowed by their Creator to change and adapt very quickly in an almost constantly changing environment. Bacteria have tremendous variability, yet there are limits to such change: the continuity, stability, and reliability of such bacteria are well known. There is a tradeoff: for survival in a clinical environment (i.e. antibiotics prevalent), there is a metabolic cost in terms of slower growth. This phenomenon is called antagonistic pleiotropy. Antagonistic pleiotropy refers to the genetic expression of multiple competing effects, some beneficial, but others harmful to the organism. It often involves a case where reproduction and viability counter each other in biological fitness (Gillen and Anderson 2008). The gene provides a benefit in one circumstance, but has cost in another. A conclusion of these experiments is that acquiring antibiotic resistance through mutation or horizontal transfer is costly.
So, is CA-MRSA really new and evolving? Yes and no. The specific strain, S. aureus, USA300 may be new (from about 1997); however, pathogenic strains that are similar to it are probably not new. S. aureus has probably undergone significant change since its creation at the time of Genesis 1. God made all life (including S. aureus) to adapt to changing conditions. Therefore, variation through time is the norm in microbes and all other creatures. In this fallen world, all creatures can cause problems for other creatures. S. aureus were made to adapt to a changing world. In order to survive, they have mutated, changed, and adapted (although they remain the same basic kind of bacteria). Sometimes in the process of changing, they cause disease in their host organism. They do not change to harm their hosts, but they sometimes do because they have themselves become subject to the Curse’s law of decay and corruption, as have their hosts.
Already-existing pathogenic genes, not novel genes, are transferred to residential skin or nasal S. aureus to produce a new strain (fig. 11). The mechanisms are not innovative and are limited in nature. They are not novel in nature; these genes are new to S. aureus only. CA-MRSA is more aggressive, virulent, and dangerous than the historical Staph infection (methicillin sensitive S. aureus).
The change in MRSA is occurring, and as it could possibly become more dangerous, more research is needed on the mechanisms of transfer and change in the new strains of MRSA to confront this threat. Is the ping-pong phenomenon, the new variants of S. aureus an example of evolution in action? If one uses the term evolution to simply mean change, yes. If one is referring to a larger, upward onward evolution descent with modification, no.
The mechanisms responsible for the change required producing limited descent with modification: chiefly natural selection acting on random variations or mutations. The changes seen in S. aureus are primarily due to a series of horizontal transfers and adaptations. There is variation to meet the demand of the new environment. One can readily observe changes like these in other pathogens and parasites. Many bacteria like E. coli, Serratia marcescens, and Mycobacterium tuberculosis do change over time. Bacteria are master adapters. The ping-pong phenomena might be described microevolution, variation, or horizontal evolution, but none of the changes lead to upward, onward Darwinian macroevolution. The new CA-MRSA strains are still Staphylococcus aureus. Regardless of the language, knowledge of emerging CA-MRSA is medically important for those combatting infectious diseases.
Definitions
Species
In microbiology, a species is one of the basic units of classification and taxonomic rank. For our purposes here, we define Staphylococcus species in the way Bergey’s Systematic Manual of Bacteriology (Garrity 2005) does.
Epivar
This is the definition of the second term.
Strains
A strain is a genetic variant or subtype of a bacterium. There are about 1100 strains of MRSA.
Biotypes
Strains that have specific profile such as antibiotic, or a molecular.
Antibiogram
An antibiogram is the result of a laboratory testing for the sensitivity of an isolated bacteria strain or biotype to different antibiotics. It is by definition the outcome of a Kirby-Bauer disk diffusion test.
Antibiotypes
Strains that have specific profile such as antibiotic sensitivity test.
Ciprofloxacin (Cipro)
An antibiotic that can treat a number of bacterial infections. It is a second-generation fluoroquinolone.
MRSA carriage (colonization)
Different from MRSA invasive disease (an infection).
References
Alam M. T., T. D. Read, R. A. Petit III, S. Boyle-Vavra, L. G. Miller, S. J. Eells, R. S. Daum, and M. Z. David. 2015. “Transmission and Microevolution of USA300 MRSA in U.S. Households: Evidence From Whole-Genome Sequencing.” mBio 6 (2): e00054-15. doi:10.1128/mBio.00054-15.
Anderson, K. 2005. “Is Bacterial Resistance to Antibiotics an Appropriate Example of Evolutionary Change?” Creation Research Society Quarterly 41 (4): 318–26.
Courvalin, P., R. LeClercq, and L. B. Rice. eds. 2011. Antibiogram. Washington DC: ASM Press.
Ender, M., N. McCallum, R. Adhikari, and B. Berger-Bächi. 2004. “Fitness Cost of SCCmec and Methicillin Resistance Levels in Staphylococcus aureus.” Antimicrobial Agents and Chemotherapy 48 (6): 2295–97.
Fox, J. ed. 2015. The Threat of MRSA. Washington DC: American Academy of Microbiology.
Garrity, G. M. (ed.) 2005. Bergey’s Manual of Systematic Bacteriology, Vol 2. The Proteobacteria, Part C. 2nd ed. Berlin, Germany: Springer-Verlag.
Gillen, A. 2009. “The Genesis of Methicillin-Resistant Staphylococcus aureus.” https://answersingenesis.org/natural-selection/antibiotic-resistance/the-genesis-of-methicillin-resistant-staphylococcus-aureus/.
Gillen, A. L. 2014. “The Wondrous Weaving of the Skin and its Microbiome.” https://answersingenesis.org/human-body/wonderfully-made-design-skin-and-its-microbiome/.
Gillen, A. L. 2015. The Genesis of Germs: Disease and the Coming Plagues in a Fallen World. Green Forest, Arkansas: Master Books.
Gillen, A. L. and S. Anderson. 2008. “Darwin at the Drugstore? Testing the Biological Fitness of Antibiotic Resistant Bacteria.” https://answersingenesis.org/natural-selection/antibiotic-resistance/darwin-at-the-drugstore/.
Gillen, A. L., W. O. Daycock, and A. Serafin. 2014. “High MRSA Carriage Rate among Nursing Microbiology Students.” Advances in Microbiology 4: 871–77. doi:10.4236/aim.2014.413096.
Haddad, L. E. and S. Moineau. 2013. “Characterization of a Novel Panton-Valentine Leukocidin (PVL)-Encoding Staphylococcal Phage and Its Naturally PVL-Lacking Variant.” Applied and Environmental Microbiology 79 (8): 2828–32.
Mitchell, C. R. 2015. “Methicillin-resistant Staphylococcus aureus: Mechanisms of Virulence and Antibiotic Sensitivity.” Senior Honors Thesis. Lynchburg, Virginia: Liberty University.
Purdom, G. and K. L. Anderson. 2008. “A Creationist Perspective of Beneficial Mutation in Bacteria.” In Proceedings of the Sixth International Conference on Creationism. Edited by A. A. Snelling, 73–85. Pittsburgh, Pennsylvania: Creation Science Fellowship and Dallas, Texas: Institute for Creation Research. https://answersingenesis.org/genetics/mutations/a-creationist-perspective-of-beneficial-mutations-in-bacteria/.
Seifert, H. S., and V. J. DiRita. eds. 2006. Evolution of Microbial Pathogens. Washington, DC: ASM Press.
Sheeran, A. M. 2014. “Prevalence of MRSA Carriers in Dormitories at Liberty University.” Second Annual Undergraduate Research Symposium. April 12, 2014. Lynchburg, Virginia: Liberty University.
Tortora, G. J., B. R. Funke, and C. L. Case. 2015. Microbiology: An Introduction. 12th ed. San Francisco, California: Pearson Benjamin/Cummings Pub. Co.
Weigelt, J. A. ed. 2008. MRSA. New York, New York: Informa Healthcare USA, Inc.
Wim, W. and A. J. Neeling. 2005. “Successful Search-and- Destroy Policy for Methicillin-Resistant Staphylococcus aureus in The Netherlands: Response.” Journal of Clinical Microbiology 43 (4): 2034–35. doi:10.1128/JCM.43.4.2034-2035.2005.
Appendix
List of Main Antibiotics Used in Kirby-Bauer Test Antibiotic/Drug Dosage in Kirby-Bauer Test
- Ciprofloxacin 5
- Clindamycin 2
- Erythromycin E-15
- Doxycycline D-30
- Vancomycin VA-30
- Mupirocin M 20